UAB Condor Pilot
https://docs.rc.uab.edu/
Please use the new documentation url https://docs.rc.uab.edu/ for all Research Computing documentation needs.
As a result of this move, we have deprecated use of this wiki for documentation. We are providing read-only access to the content to facilitate migration of bookmarks and to serve as an historical record. All content updates should be made at the new documentation site. The original wiki will not receive further updates.
Thank you,
The Research Computing Team
The UAB Condor Pilot explored the utility of the cloud computing paradigm to research applications using aggregated, unused compute cycles harvested from many computers. The pilot established a demonstration spare-cycle compute fabric using the Condor scheduler and compared the performance of a molecular docking workflow on this fabric and several larger production Condor fabrics to the performance of the same workflow running on our campus compute cluster Cheaha.
The UAB Condor Pilot successfully demonstrated the value of harvested unused compute cycles to the molecular docking workflow. The results suggest that similar applications, especially those that can scale by repeating the same task on distinct data sets, will likewise benefit from the abundant compute resources that can be harvested via a Condor compute fabric. The UAB Condor Pilot ended in May 2012 with the presentation of our results (pdf) at Condor Week 2012. This page is available as a PDF.
Background
Condor
Condor is a resource allocation and management system designed to simplify harvesting idle compute cycles from under-utilized computers. Condor is a production-quality software system developed by researchers at the University of Wisconsin. It is deployed in a wide range of environments from lab or departmental compute pools with 10's of processors to global compute fabrics such as the Open Science Grid (OSG) that averages between 80,000 to 100,000 processors.
Condor organizes computers offering their spare cycles into resource collections called "pools". A Condor pool provides compute cycles to applications in the context of jobs by matching criteria expressed by the applications to capabilities expressed by the resources; a process descriptively entitled "match making". A special focus of Condor is to avoid inconveniencing resource providers who offer their spare cycles to applications. Condor can quickly evacuate a job from an assigned resource, e.g. an end user's desktop computer, should activity on the mouse or keyboard be detected. Condor also provides mechanisms to the job owner to checkpoint work and avoid loss of forward progress in this event. Condor is a flexible system that can harness loosely-coupled systems as well as traditional tightly-coupled cluster systems. The focus of this pilot was on harvesting compute cycles of loosely coupled systems.
There is an active user and developer community around Condor. Condor is supported on Linux, Mac and Windows ensuring utility to a broad spectrum of applications, from scientific computations to large scale statistical analyses. There are personal instances to support individuals migrating or developing their own workflows. The loosely-coupled nature of Condor resource collections make it straight forward to dynamically scale out on popular cloud computing fabrics such as Amazon EC2.
Molecular Docking
Molecular docking is a process for discovering an ideal orientation between two molecules: the receptor and the ligand. There are a number of approaches that can be taken to explore how a ligand (drug) can bind to a receptor (protein). The approach used in this pilot was a conformational space search using genetic algorithms which evolve the orientation of the molecules to find the most likely orientation to support docking of the molecules, as implemented by the open source AutoDock application from The Scripps Research Institute.
This virtual screening of protein-drug interactions is computationally intense and can incorporate large databases of chemical compounds. This makes it an ideal candidate for leveraging as many compute resources as possible to during the screening process. Furthermore, the structure of this workflow using AutoDock analyzes each receptor-ligand pair independently, making it an ideal candidate for leveraging the loosely couple collection of computers made available through the Condor system.
Molecular docking is a virtual exploration of molecules made possible by a physical-world workflow that involves discovery of the molecular structure of proteins via X-ray crystallography. This process is nicely described by this video documenting X-ray crystallography at the Institute of Molecular and Cell Biology in Strasbourg, France. Similar facilities at Argonne National Laboratory are used by researchers at UAB's Center for Biophysical Sciences and Engineering to explore the structure of proteins.
Components of Pilot
Condor Testbeds
The pilot leveraged three representative Condor implementations to assess the performance of the molecular docking workflow. The first testbed was a demonstration UAB campus Condor pool established as part of the pilot. This pool included approximately 40 64bit Linux workstations from labs in the Department of Computer and Information Sciences and 40 32bit Windows desktops from UAB IT Desktop Support. These systems are representative of computers on campus that typically have long idle periods and which could offer their cycles to compute tasks which need them.
The second testbed was the University of Wisconsin's campus Condor pool. This is a production Condor pool that has over 1000 64bit Linux workstations. This pool is operated by the Center for High-Throughput Computing (CHTC) at the University of Wisconsin and is part of the production compute fabric available to researchers at the University of Wisconsin. It was made available to us as part of our pilot through the generous support of CHTC and Dr. Miron Livny.
The third testbed was a dynamically provisioned Condor pool made available by the Engage VO (Virtual Organization). Engage VO leverages compute cycles provided by the Open Science Grid (OSG). The Engage VO was established to encourage exploration of OSG by users new to grid computing environments. It operates a dynamically provisioned Condor pool using a technology called glideinWMS to affiliate available compute cycles in OSG with a virtual Condor pool. This is the essence of the on-demand resource allocation of cloud computing. The Engage VO is operated by RENCI (RENaissance Computing Institute) a multi-institutional research resource for North Carolina. Engage VO access and support was provided by John McGee, PI of the grant that established the Engage VO.
Autodock Workflow
The UAB Condor Pilot migrated a representative molecular docking workflow that runs on UAB's 888 core campus compute cluster using the SGE batch scheduler to a workflow that could be executed in a Condor scheduling and resource allocation environment. This workflow is built on AutoDock molecular docking application from The Scripps Research Institute. Standard AutoDock workflows treat each receptor-ligand docking search as an independent process. A large database of ligands and receptors is broken down into individual docking pairs each of which are processed by an independent instance of the AutoDock application. This independence between work units makes an AutoDock-based molecular docking workflow an inherently good candidate for migration to the distributed and dynamic compute fabrics typical of Condor environments.
The primary consideration in migrating this workflow to Condor was to re-package the data sets so they could be effectively distributed to many unrelated compute nodes. In traditional compute clusters all nodes are in close physical proximity and operated as a common administrative unit. This makes it possible to provide access to a shared file system across the cluster and hide data distribution to compute nodes behind a global file name space and high speed networks. In a Condor environment, compute nodes are typically spread across many physical locations and their administrative independence makes it more difficult to provide a global file name space. The potential variability in network speeds to compute nodes also necessitates disciplined data transfer. It is detrimental to distribute the full data set to all compute nodes when each node may only work on a small portion of the data.
Given the three Condor fabrics targeted in the pilot, our focus was on a simple, static packaging that would treat one receptor and five ligands as a single job. That is, each job would compute 5 molecular pairings. Furthermore, each job would only require that the 1 target receptor and the 5 candidate ligands be staged to the compute node thus providing an adequate balance between data set size (job staging data transfer time) and computational time.
A second consideration in the workflow migration was availability of tools on the compute nodes. The standard AutoDock workflow consists of two parts: data preparation and molecular docking. The data preparation component could potentially be distributed but it has many application dependencies that would need to be resolved on each compute node. Because data preparation is computationally light-weight in comparison to the docking search effort and because it is independent of the actual docking, we chose to run this step once on a dedicated resource and produce a ready-to-run data set for the experimental molecular docking runs. The molecular docking step is performed by the AutoDock executable which is self contained and easily distributed along with the data sets.
Finally, there are syntactic differences between SGE and Condor which required conversion of the job submit code to support Condor in lieu of SGE. Given the serial-job orientation of the existing SGE-based AutoDock workflow, this was mainly an effort in translating the job submit code. While powerful and flexible, Condor has a fairly easy learning curve that facilitates adoption of basic but useful functionality first, allowing advanced features to be adopted as the user gains experience. Additionally, Condor can be run as a "personal" instance to support development of workflows on a single workstation.
Participants
The UAB Condor Pilot was a collaboration between multiple organizations on campus. It was sparked by an inquiry from Dr. Charles Prince, Assistant Vice President for Research, seeking to understand if idle desktops at UAB could contribute effectively to research computing demand. The molecular docking workflow and data sets were provided by Dr. Stephen Aller of the Department of Pharmacology and Toxicology in UAB's School of Medicine. The resources to build the demonstration campus Condor pool were provided by the Department of Computer and Information Sciences (CIS) in the College of Arts and Sciences and by UAB IT Desktop Support Services. Project management and the application development were provided by the UAB IT Research Computing group.
Performance Comparison
In order to assess the utility of idle compute cycles, performance of the molecular docking workflow was compared on four compute fabrics: the UAB 888-core campus compute cluster in use for most large-scale computational workflows today, including the molecular docking workflow explored in this pilot; the demonstration UAB Condor pool of 40 Linux workstations and 40 Windows workstations; the University of Wisconsin's production Condor pool; and the Condor pool composed of OSG resources made available by the Engage VO.
The molecular docking workflow analyzed 4 receptor proteins against a database of 5440 ligands. This data set was divided into a static collection of jobs that each included 1 receptor and 5 ligands for a total of 1088 independent analysis jobs which would need to be distributed across the available compute resources of each of the four compute fabrics listed above.
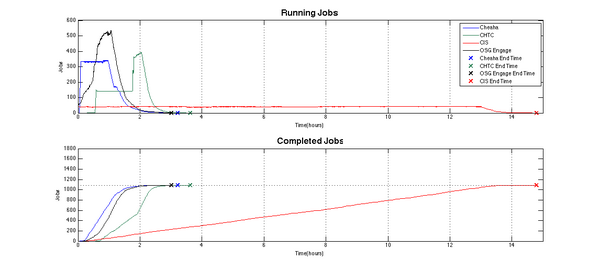
The following graphic highlights a simple comparison of run-time performance of the molecular docking workflow on each fabric. The upper graph describes the number of jobs running at any one point in time on the target compute fabric. This graph illustrates the variable nature of available compute resources, especially for the dynamic harvesting of idle cycles on the Condor-based fabrics. The lower graph records the rate of completion of jobs and illustrates the typically linear relationship between the completion rate and the number of available resources.
The UAB campus cluster Cheaha (blue lines) had approximately 300 cores available for the duration of this workflow on the day that it was run. This resulted in steady, linear progress toward workflow completion. The curved slopes at the beginning and end, represent job submission rates and workflow drain times as the remaining jobs completed. The overall runtime was approximately 3.5 hours.
The UAB Condor pilot pool (red lines) had only 40 cores available for the duration of this workflow. Only the Linux workstations where used since the Windows resources did not support the Bash control scripts initially. While the overall run-time was significantly affected by this small resource pool, it should be noted that the progress remained linear, though at a much more gentle slope. This illustrates the adaptability of the workflow structure of these serial jobs.
The Wisconsin production Condor pool (green lines) had two periods of resources availability during the run. The first period had approximately 150 idle cores available for the first hour. This jumped to close to 400 cores during the second hour. The corresponding increase in the slope of the lower job completion graph at this transition point should be noted. As more idle resources became available, the molecular docking workflow performance increased. This is an important characteristic of serial, or rather, pleasantly parallel jobs; jobs which easily translate into a parallel adaptation because they can work independently of all other jobs in the workflow. This is a very common scenario in research data process workflows. The performance of pleasantly parallel jobs is very responsive to available resources. The workflow completed in approximately 3.75 hours on the Wisconsin Condor pool.
The execution on the Engage VO Condor pool had a variable number of resources available from OSG during the run, peaking at approximately 500 cores. Nonetheless, there was steady progress toward completion and, in fact, this OSG-based execution completed in the shortest overall runtime: approximately 3 hours.
While the overall runtime of Cheaha, the Wisconsin pool, and OSG resources are roughly similar, they clearly demonstrate that idle compute cycles harnessed by Condor can perform as well as, or even better than, UAB's dedicated compute cluster for this class of compute jobs.
The original data and methods to reproduce these experimental runs and a more details analysis of results are published on the UAB Condor project site.
Conclusions
The primary conclusion discussed in the Performance Analysis above is that idle compute cycles harnessed by the Condor resource scheduler are capable of significant contributions to research computing needs at UAB. As such, UAB IT's Research Computing group recommends the adoption and support of Condor computing at UAB.
The performance summary above also demonstrates a key characteristic of high-throughput computing jobs: they scale almost linearly with the addition of compute resources. Since each task in the queue is independent of other tasks, the resource scheduler can easily assign tasks as resources become available. When combined with a flexible resource scheduler like Condor, which can harness many different compute platforms across domains, large compute pools can be assembled to address demanding workflow requirements.
The static job configuration in this test could stand improvement. While it facilitated a quick conversion of the workflow from SGE to Condor, it did lead to imbalances in effort. These can be seen as the long tails at the end of the workflow completion. These tails were caused by longer running jobs being scheduled late in the overall workflow. Allowing a granularity of 1-to-1 receptor-ligand pairs could improve the balance across compute resources. The trade-off with this approach is that some docking searches are very quick and would have more scheduling overhead than actual runtime. Additional frameworks that layer on top of Condor and address can support this model by harnessing compute resources for longer cycles and repeatedly assigning tasks as individual workers complete their tasks.
Comparing the utility of Linux and Windows resources, our pilot with molecular docking and AutoDock also showed that Linux resources are easier to use for this application than Windows workstations. This is largely a function of the application support for different platforms. Autodock is supported on Linux and Windows, but the supporting job scripts written in Bash required additional software components on Windows. This was readily resolved on UAB IT Desktop Support resources by added the Cygwin framework to the Windows workstations, however, this may not be available on all Windows platforms. A test run with this enhanced configuration found the workflow adapted easily to Cygwin but the 32bit architecture of the workstation did slow AutoDock performance. It is expected that a production deployment would include more current hardware resources and that 64bit Windows systems will be more widely available.
The ability to move this same workflow, with very little adjustment, across a pilot Condor pool, production campus pool and national grid fabric demonstrates the advantage Condor has in portability. One of the key barriers limiting research adoption of new platforms (even more powerful ones) is the effort to port the workflow to the new resource environment. With Condor, local compute resources can be used to develop a workflow (including a simple personal deployment) and then this workflow can easily be move to larger and larger compute resources as demand grows.
References
Performance improvements to molecular docking workflows using Autodock are a common topic in the literature. The following papers address various approaches that have been taken to improve molecular docking workflow performance on a wide variety of hardware platforms, from BlueGene computers to Hadoop clusters. These papers used a reference docking set to compare performance, while our pilot preferred a data set representative of existing workflows on our cluster.
- Ellingson S. et. al. High-Throughput Virtual Molecular Docking: Hadoop Implementation of Autdock4 on a Private Cloud (2011)
- Norgan A., et. al. Multilevel Parallelization of Autodock 4.2 (2011)
- Collingnon B., et. al. Task‐Parallel Message Passing Interface Implementation of Autodock4 for Docking of Very Large Databases of Compounds Using High-Performance Super-Computers (2010)
- Huang N., et. al. Benchmarking Sets for Molecular Docking (2006)