Obsolete: Resources and Obsolete: Cheaha Getting Started: Difference between pages
Test admin (talk | contribs) (Obsolete content for getting started on Resources) |
Test admin (talk | contribs) m (Test admin moved page Obsolete Cheaha Getting Started to Obsolete: Cheaha Getting Started: Consistency with colon separator) |
||
| Line 1: | Line 1: | ||
Cheaha is a cluster computing environment for UAB researchers. Information about the history and future plans for Cheaha is available on the [[Cheaha]] page. | |||
== Access (Cluster Account Request) == | |||
To request an account on [[Cheaha]], please {{CheahaAccountRequest}}. Please include some background information about the work you plan on doing on the cluster and the group you work with, ie. your lab or affiliation. | |||
'''NOTE:''' | |||
The email you send to support@listserv.uab.edu will send back a '''confirmation email which must be acknowledged''' in order to submit the account request. | |||
This additional step is meant to cut down on spam to the support list and is only needed for the initial account creation request sent to the support list. | |||
Usage of Cheaha is governed by [https://www.uab.edu/policies/content/Pages/UAB-IT-POL-0000004.aspx UAB's Acceptable Use Policy (AUP)] for computer resources. | |||
=== External Collaborator=== | |||
To request an account for an external collaborator, please follow the steps [https://docs.uabgrid.uab.edu/wiki/Collaborator_Account here.] | |||
== Login == | |||
===Overview=== | |||
Once your account has been created, you'll receive an email containing your user ID, generally your Blazer ID. You can [https://rc.uab.edu log into Cheaha via your web browser] using the new web-based HPC experience. | |||
You can also log into Cheaha via a traditional SSH client. Most UAB Windows workstations already have an SSH client installed, possibly named '''SSH Secure Shell Client''' or [http://www.chiark.greenend.org.uk/~sgtatham/putty/ PuTTY]. Linux and Mac OS X systems should have an SSH client installed by default. | |||
Usage of Cheaha is governed by [https://www.uab.edu/policies/content/Pages/UAB-IT-POL-0000004.aspx UAB's Acceptable Use Policy (AUP)] for computer and network resources. | |||
===Client Configuration=== | |||
This section will cover steps to configure Windows, Linux and Mac OS X clients to connect to Cheaha. | |||
The official DNS name of Cheaha's frontend machine is ''cheaha.rc.uab.edu''. If you want to refer to the machine as ''cheaha'', you'll have to either add the "rc.uab.edu" to you computer's DNS search path. On Unix-derived systems (Linux, Mac) you can edit your computers /etc/resolv.conf as follows (you'll need administrator access to edit this file) | |||
<pre> | |||
search rc.uab.edu | |||
</pre> | |||
Or you can customize your SSH configuration to use the short name "cheaha" as a connection name. On systems using OpenSSH you can add the following to your ~/.ssh/config file | |||
<pre> | |||
Host cheaha | |||
Hostname cheaha.rc.uab.edu | |||
</pre> | |||
==== | ====Linux==== | ||
Linux systems, regardless of the flavor (RedHat, SuSE, Ubuntu, etc...), should already have an SSH client on the system as part of the default install. | |||
# Start a terminal (on RedHat click Applications -> Accessories -> Terminal, on Ubuntu Ctrl+Alt+T) | |||
# At the prompt, enter the following command to connect to Cheaha ('''Replace blazerid with your Cheaha userid''') | |||
ssh '''blazerid'''@cheaha.rc.uab.edu | |||
====Mac OS X==== | |||
Mac OS X is a Unix operating system (BSD) and has a built in ssh client. | |||
# Start a terminal (click Finder, type Terminal and double click on Terminal under the Applications category) | |||
# At the prompt, enter the following command to connect to Cheaha ('''Replace blazerid with your Cheaha userid''') | |||
ssh '''blazerid'''@cheaha.rc.uab.edu | |||
=== | ====Windows==== | ||
There are many SSH clients available for Windows, some commercial and some that are free (GPL). This section will cover two clients that are commonly found on UAB Windows systems. | |||
=====MobaXterm===== | |||
[http://mobaxterm.mobatek.net/ MobaXterm] is a free (also available for a price in a Profession version) suite of SSH tools. Of the Windows clients we've used, MobaXterm is the easiest to use and feature complete. [http://mobaxterm.mobatek.net/features.html Features] include (but not limited to): | |||
* SSH client (in a handy web browser like tabbed interface) | |||
* Embedded Cygwin (which allows Windows users to run many Linux commands like grep, rsync, sed) | |||
* Remote file system browser (graphical SFTP) | |||
* X11 forwarding for remotely displaying graphical content from Cheaha | |||
* Installs without requiring Windows Administrator rights | |||
== | Start MobaXterm and click the Session toolbar button (top left). Click SSH for the session type, enter the following information and click OK. Once finished, double click cheaha.rc.uab.edu in the list of Saved sessions under PuTTY sessions: | ||
{| border="1" cellpadding="5" | |||
!Field | |||
!Cheaha Settings | |||
|- | |||
|'''Remote host''' | |||
|cheaha.rc.uab.edu | |||
|- | |||
|'''Port''' | |||
|22 | |||
|- | |||
|} | |||
=====PuTTY===== | |||
[http://www.chiark.greenend.org.uk/~sgtatham/putty/ PuTTY] is a free suite of SSH and telnet tools written and maintained by [http://www.pobox.com/~anakin/ Simon Tatham]. PuTTY supports SSH, secure FTP (SFTP), and X forwarding (XTERM) among other tools. | |||
* Start PuTTY (Click START -> All Programs -> PuTTY -> PuTTY). The 'PuTTY Configuration' window will open | |||
* Use these settings for each of the clusters that you would like to configure | |||
{| border="1" cellpadding="5" | |||
!Field | |||
!Cheaha Settings | |||
|- | |||
|'''Host Name (or IP address)''' | |||
|cheaha.rc.uab.edu | |||
|- | |||
|'''Port''' | |||
|22 | |||
|- | |||
|'''Protocol''' | |||
|SSH | |||
|- | |||
|'''Saved Sessions''' | |||
|cheaha.rc.uab.edu | |||
|- | |||
|} | |||
* Click '''Save''' to save the configuration, repeat the previous steps for the other clusters | |||
* The next time you start PuTTY, simply double click on the cluster name under the 'Saved Sessions' list | |||
==== | =====SSH Secure Shell Client===== | ||
''' | SSH Secure Shell is a commercial application that is installed on many Windows workstations on campus and can be configured as follows: | ||
* Start the program (Click START -> All Programs -> SSH Secure Shell -> Secure Shell Client). The 'default - SSH Secure Shell' window will open | |||
* Click File -> Profiles -> Add Profile to open the 'Add Profile' window | |||
* Type in the name of the cluster (for example: cheaha) in the field and click 'Add to Profiles' | |||
* Click File -> Profiles -> Edit Profiles to open the 'Profiles' window | |||
* Single click on your new profile name | |||
* Use these settings for the clusters | |||
{| border="1" cellpadding="5" | |||
!Field | |||
!Cheaha Settings | |||
|- | |||
|'''Host name''' | |||
|cheaha.rc.uab.edu | |||
|- | |||
|'''User name''' | |||
|blazerid (insert your blazerid here) | |||
|- | |||
|'''Port''' | |||
|22 | |||
|- | |||
|'''Protocol''' | |||
|SSH | |||
|- | |||
|'''Encryption algorithm''' | |||
|<Default> | |||
|- | |||
|'''MAC algorithm''' | |||
|<Default> | |||
|- | |||
|'''Compression''' | |||
|<None> | |||
|- | |||
|'''Terminal answerback''' | |||
|vt100 | |||
|- | |||
|} | |||
* Leave 'Connect through firewall' and 'Request tunnels only' unchecked | |||
* Click '''OK''' to save the configuration, repeat the previous steps for the other clusters | |||
* The next time you start SSH Secure Shell, click 'Profiles' and click the cluster name | |||
=== | === Logging in to Cheaha === | ||
No matter which client you use to connect to the Cheaha, the first time you connect, the SSH client should display a message asking if you would like to import the hosts public key. Answer '''Yes''' to this question. | |||
* Connect to Cheaha using one of the methods listed above | |||
* Answer '''Yes''' to import the cluster's public key | |||
** Enter your BlazerID password | |||
* After successfully logging in for the first time, You may see the following message '''just press ENTER for the next three prompts, don't type any passphrases!''' | |||
It doesn't appear that you have set up your ssh key. | |||
This process will make the files: | |||
/home/joeuser/.ssh/id_rsa.pub | |||
/home/joeuser/.ssh/id_rsa | |||
/home/joeuser/.ssh/authorized_keys | |||
Generating public/private rsa key pair. | |||
Enter file in which to save the key (/home/joeuser/.ssh/id_rsa): | |||
** Enter file in which to save the key (/home/joeuser/.ssh/id_rsa):'''Press Enter''' | |||
** Enter passphrase (empty for no passphrase):'''Press Enter''' | |||
** Enter same passphrase again:'''Press Enter''' | |||
Your identification has been saved in /home/joeuser/.ssh/id_rsa. | |||
Your public key has been saved in /home/joeuser/.ssh/id_rsa.pub. | |||
The key fingerprint is: | |||
f6:xx:xx:xx:xx:dd:9a:79:7b:83:xx:f9:d7:a7:d6:27 joeuser@cheaha.rc.uab.edu | |||
==== Users without a blazerid (collaborators from other universities) ==== | |||
** If you were issued a temporary password, enter it (Passwords are CaSE SensitivE!!!) You should see a message similar to this | |||
You are required to change your password immediately (password aged) | |||
WARNING: Your password has expired. | |||
You must change your password now and login again! | |||
Changing password for user joeuser. | |||
Changing password for joeuser | |||
(current) UNIX password: | |||
*** (current) UNIX password: '''Enter your temporary password at this prompt and press enter''' | |||
*** New UNIX password: '''Enter your new strong password and press enter''' | |||
*** Retype new UNIX password: '''Enter your new strong password again and press enter''' | |||
*** After you enter your new password for the second time and press enter, the shell may exit automatically. If it doesn't, type exit and press enter | |||
*** Log in again, this time use your new password | |||
Congratulations, you should now have a command prompt and be ready to start [[Cheaha_GettingStarted#Example_Batch_Job_Script | submitting jobs]]!!! | |||
== Hardware == | |||
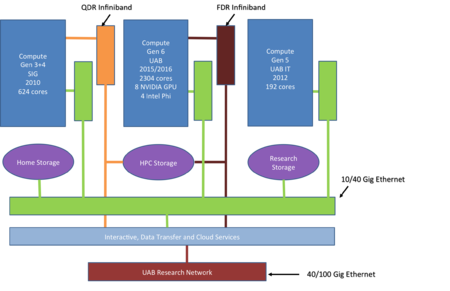
[[Image:Chehah2_2016.png|center|thumb|450px|Logical Diagram of Cheaha Configuration]] | |||
The Cheaha Compute Platform includes commodity compute hardware, totaling 2800 compute cores and over 4.7PB of usable storage (6.6PB raw capacity). The following descriptions highlight the current hardware profile that provides an aggregate theoretical peak performance of 468 teraflops. | |||
* Compute | |||
** 36 Compute Nodes with two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 128GB DDR4 RAM, FDR InfiniBand and 10GigE network cards | |||
** 38 Compute Nodes with two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 256GB DDR4 RAM, FDR InfiniBand and 10GigE network cards | |||
** 14 Compute Nodes with two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 384GB DDR4 RAM, FDR InfiniBand and 10GigE network cards | |||
** 4 Compute Nodes with Nvidia Tesla K80 and two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 128GB DDR4 RAM, FDR InfiniBand and 10GigE network cards | |||
** 4 Compute Nodes with Intel Phi coprocessor SE10/7120 and two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 128GB DDR4 RAM, FDR InfiniBand and 10GigE network cards | |||
** 18 Compute Nodes with two 14 core processors (Intel Xeon E5-2680 v4 2.4GHz)with 256GB DDR4 RAM, four NVIDIA Tesla P100 16GB GPUs, EDR InfiniBand and 10GigE network cards | |||
* Networking | |||
**FDR and EDR InfiniBand Switch | |||
** 10Gigabit Ethernet Switch | |||
* Storage -- DDN SFA12KX with GPFS) | |||
** 2 x 12KX40D-56IB controllers | |||
** 10 x SS8460 disk enclosures | |||
** 825 x 4K SAS drives | |||
* Management | |||
** Management node and gigabit switch for cluster management | |||
** Bright Advanced Cluster Management software licenses | |||
== Cluster Software == | |||
* BrightCM 7.2 | |||
* CentOS 7.2 x86_64 | |||
* [[Slurm]] 15.08 | |||
== Queuing System == | |||
All work on Cheaha must be submitted to '''our queuing system ([[Slurm]])'''. A common mistake made by new users is to run 'jobs' on the login node. This section gives a basic overview of what a queuing system is and why we use it. | |||
=== What is a queuing system? === | |||
* Software that gives users fair allocation of the cluster's resources | |||
* Schedules jobs based using resource requests (the following are commonly requested resources, there are many more that are available) | |||
** Number of processors (often referred to as "slots") | |||
** Maximum memory (RAM) required per slot | |||
** Maximum run time | |||
* Common queuing systems: | |||
** '''[[Slurm]]''' | |||
** Sun Grid Engine (Also know as SGE, OGE, GE) | |||
** OpenPBS | |||
** Torque | |||
** LSF (load sharing facility) | |||
[http://slurm.schedmd.com/ Slurm] is a queue management system and stands for Simple Linux Utility for Resource Management. Slurm was developed at the Lawrence Livermore National Lab and currently runs some of the largest compute clusters in the world. '''[[Slurm]]''' is now the primary job manager on Cheaha, it replaces SUN Grid Engine ([[https://docs.uabgrid.uab.edu/wiki/Cheaha_GettingStarted_deprecated SGE]]) the job manager used earlier. Instructions of using SLURM and writing SLURM scripts for jobs submission on Cheaha can be found '''[[Slurm | here]]'''. | |||
=== Typical Workflow === | |||
* Stage data to $USER_SCRATCH (your scratch directory) | |||
* Research how to run your code in "batch" mode. Batch mode typically means the ability to run it from the command line without requiring any interaction from the user. | |||
* Identify the appropriate resources needed to run the job. The following are mandatory resource requests for all jobs on Cheaha | |||
** Maximum memory (RAM) required per slot | |||
** Maximum runtime | |||
* Write a job script specifying queuing system parameters, resource requests and commands to run program | |||
* Submit script to queuing system (sbatch script.job) | |||
* Monitor job (squeue) | |||
* Review the results and resubmit as necessary | |||
* Clean up the scratch directory by moving or deleting the data off of the cluster | |||
=== Resource Requests === | |||
Accurate resource requests are extremely important to the health of the over all cluster. In order for Cheaha to operate properly, the queing system must know how much runtime and RAM each job will need. | |||
==== Mandatory Resource Requests ==== | |||
* -t, --time=<time> | |||
Set a limit on the total run time of the job allocation. If the requested time limit exceeds the partition's time limit, the job will be left in a PENDING state (possibly indefinitely). | |||
* For Array jobs, this represents the maximum run time for each task | |||
** For serial or parallel jobs, this represents the maximum run time for the entire job | |||
* --mem-per-cpu=<MB> | |||
Mimimum memory required per allocated CPU in MegaBytes. | |||
==== Other Common Resource Requests ==== | |||
* -N, --nodes=<minnodes[-maxnodes]> | |||
Request that a minimum of minnodes nodes be allocated to this job. A maximum node count may also be specified with maxnodes. If only one number is specified, this is used as both the minimum and maximum node count. | |||
* -n, --ntasks=<number> | |||
sbatch does not launch tasks, it requests an allocation of resources and submits a batch script. This option advises the Slurm controller that job steps run within the allocation will launch a maximum of number tasks and to provide for sufficient resources. The default is one task per node | |||
* --mem=<MB> | |||
Specify the real memory required per node in MegaBytes. | |||
* -c, --cpus-per-task=<ncpus> | |||
Advise the Slurm controller that ensuing job steps will require ncpus number of processors per task. Without this option, the controller will just try to allocate one processor per task. | |||
* -p, --partition=<partition_names> | |||
Request a specific partition for the resource allocation. Available partitions are: express(max 2 hrs), short(max 12 hrs), medium(max 50 hrs), long(max 150 hrs), sinteractive(0-2 hrs) | |||
=== Submitting Jobs === | |||
Batch Jobs are submitted on Cheaha by using the "sbatch" command. The full manual for sbtach is available by running the following command | |||
man sbatch | |||
==== Job Script File Format ==== | |||
To submit a job to the queuing systems, you will first define your job in a script (a text file) and then submit that script to the queuing system. | |||
The script file needs to be '''formatted as a UNIX file''', not a Windows or Mac text file. In geek speak, this means that the end of line (EOL) character should be a line feed (LF) rather than a carriage return line feed (CRLF) for Windows or carriage return (CR) for Mac. | |||
If you submit a job script formatted as a Windows or Mac text file, your job will likely fail with misleading messages, for example that the path specified does not exist. | |||
Windows '''Notepad''' does not have the ability to save files using the UNIX file format. Do NOT use Notepad to create files intended for use on the clusters. Instead use one of the alternative text editors listed in the following section. | |||
===== Converting Files to UNIX Format ===== | |||
====== Dos2Unix Method ====== | |||
The lines below that begin with $ are commands, the $ represents the command prompt and should not be typed! | |||
The dos2unix program can be used to convert Windows text files to UNIX files with a simple command. After you have copied the file to your home directory on the cluster, you can identify that the file is a Windows file by executing the following (Windows uses CR LF as the line terminator, where UNIX uses only LF and Mac uses only CR): | |||
<pre> | |||
$ file testfile.txt | |||
testfile.txt: ASCII text, with CRLF line terminators | |||
</pre> | |||
Now, convert the file to UNIX | |||
<pre> | |||
$ dos2unix testfile.txt | |||
dos2unix: converting file testfile.txt to UNIX format ... | |||
</pre> | |||
Verify the conversion using the file command | |||
<pre> | |||
$ file testfile.txt | |||
testfile.txt: ASCII text | |||
</pre> | |||
====== Alternative Windows Text Editors ====== | |||
There are many good text editors available for Windows that have the capability to save files using the UNIX file format. Here are a few: | |||
* [[http://www.geany.org/ Geany]] is an excellent free text editor for Windows and Linux that supports Windows, UNIX and Mac file formats, syntax highlighting and many programming features. To convert from Windows to UNIX click '''Document''' click '''Set Line Endings''' and then '''Convert and Set to LF (Unix)''' | |||
* [[http://notepad-plus.sourceforge.net/uk/site.htm Notepad++]] is a great free Windows text editor that supports Windows, UNIX and Mac file formats, syntax highlighting and many programming features. To convert from Windows to UNIX click '''Format''' and then click '''Convert to UNIX Format''' | |||
* [[http://www.textpad.com/ TextPad]] is another excellent Windows text editor. TextPad is not free, however. | |||
==== Example Batch Job Script ==== | |||
A shared cluster environment like Cheaha uses a job scheduler to run tasks on the cluster to provide optimal resource sharing among users. Cheaha uses a job scheduling system call Slurm to schedule and manage jobs. A user needs to tell Slurm about resource requirements (e.g. CPU, memory) so that it can schedule jobs effectively. These resource requirements along with actual application code can be specified in a single file commonly referred as 'Job Script/File'. Following is a simple job script that prints job number and hostname. | |||
'''Note:'''Jobs '''must request''' the appropriate partition (ex: ''--partition=short'') to satisfy the jobs resource request (maximum runtime, number of compute nodes, etc...) | |||
<pre> | |||
#!/bin/bash | |||
# | |||
#SBATCH --job-name=test | |||
#SBATCH --output=res.txt | |||
#SBATCH --ntasks=1 | |||
#SBATCH --partition=express | |||
#SBATCH --time=10:00 | |||
#SBATCH --mem-per-cpu=100 | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=YOUR_EMAIL_ADDRESS | |||
srun hostname | |||
srun sleep 60 | |||
</pre> | |||
Lines starting with '#SBATCH' have a special meaning in the Slurm world. Slurm specific configuration options are specified after the '#SBATCH' characters. Above configuration options are useful for most job scripts and for additional configuration options refer to Slurm commands manual. A job script is submitted to the cluster using Slurm specific commands. There are many commands available, but following three commands are the most common: | |||
* sbatch - to submit job | |||
* scancel - to delete job | |||
* squeue - to view job status | |||
We can submit above job script using sbatch command: | |||
<pre> | |||
$ sbatch HelloCheaha.sh | |||
Submitted batch job 52707 | |||
</pre> | |||
When the job script is submitted, Slurm queues it up and assigns it a job number (e.g. 52707 in above example). The job number is available inside job script using environment variable $JOB_ID. This variable can be used inside job script to create job related directory structure or file names. | |||
=== Interactive Resources === | |||
Login Node (the host that you connected to when you setup the SSH connection to Cheaha) is supposed to be used for submitting jobs and/or lighter prep work required for the job scripts. '''Do not run heavy computations on the login node'''. If you have a heavier workload to prepare for a batch job (eg. compiling code or other manipulations of data) or your compute application requires interactive control, you should request a dedicated interactive node for this work. | |||
Interactive resources are requested by submitting an "interactive" job to the scheduler. Interactive jobs will provide you a command line on a compute resource that you can use just like you would the command line on the login node. The difference is that the scheduler has dedicated the requested resources to your job and you can run your interactive commands without having to worry about impacting other users on the login node. | |||
Interactive jobs, that can be run on command line, are requested with the '''srun''' command. | |||
<pre> | |||
srun --ntasks=1 --cpus-per-task=4 --mem-per-cpu=4096 --time=08:00:00 --partition=medium --job-name=JOB_NAME --pty /bin/bash | |||
</pre> | |||
This command requests for 4 cores (--cpus-per-task) for a single task (--ntasks) with each cpu requesting size 4GB of RAM (--mem-per-cpu) for 8 hrs (--time). | |||
More advanced interactive scenarios to support graphical applications are available using [https://docs.uabgrid.uab.edu/wiki/Setting_Up_VNC_Session VNC] or X11 tunneling [http://www.uab.edu/it/software X-Win32 2014 for Windows] | |||
Interactive jobs that requires running a graphical application, are requested with the '''sinteractive''' command, via '''Terminal''' on your VNC window. | |||
<pre> | |||
sinteractive --ntasks=1 --cpus-per-task=4 --mem-per-cpu=4096 --time=08:00:00 --partition=medium --job-name=JOB_NAME | |||
</pre> | |||
Please note, sinteractive starts your shell in a screen session. Screen is a terminal emulator that is designed to make it possible to detach and reattach a session. This feature can mostly be ignored. If you application uses `ctrl-a` as a special command sequence (e.g. Emacs), however, you may find the application doesn't receive this special character. When using screen, you need to type `ctrl-a a` (ctrl-a followed by a single "a" key press) to send a ctrl-a to your application. Screen uses ctrl-a as it's own command character, so this special sequence issues the command to screen to "send ctrl-a to my app". Learn more about [https://www.gnu.org/software/screen/manual/html_node/Overview.html#Overview screen from it's documentation]. | |||
== Storage == | |||
=== Privacy === | |||
{{SensitiveInformation}} | |||
=== File and Directory Permissions === | |||
The default permissions for all user data storage locations described below are as follows. In these descriptions, the "$USER" variable should be replaced with the user's account name string: | |||
* /home/$USER - the owner ($USER) of the directory can read, write/delete, and list files. No other users or groups have permissions to this directory. | |||
* /data/user/$USER - the owner ($USER) of the directory can read, write/delete, and list files. No other users or groups have permissions to this directory. | |||
* /scratch/$USER - the owner ($USER) of the directory can read, write/delete, and list files. No other users or groups have permissions to this directory. | |||
* /data/projects/<projectname> - a PI can request project space for their lab or specific collaborations. The project directory is created with the PI/requestor as the user-owner and a dedicated collaboration group as the group-owner. The PI and all members of the dedicated collaboration group have can read, write/delete, and list files. No privileges are granted to other users of the system. Additional controls can be implemented via access control lists (ACLs). The PI/requestor can modify the ACLs to allow additional access to specific users. | |||
These permissions are the default configuration. While it is possible to modify these permissions or change the group owner of a file to any group to which a user belongs, users are encouraged to work within the default configuration and contact support@listserv.uab.edu if the default permissions are not adequate. Setting up a collaboration group and associated project directory can address most collaboration need while keep data access restricted to the minimum necessary users for the collaboration. | |||
Additional background on Linux file system permissions can be found here: | |||
* https://its.unc.edu/research-computing/techdocs/how-to-use-unix-and-linux-file-permissions/ | |||
* https://www.rc.fas.harvard.edu/resources/documentation/linux/unix-permissions/ | |||
* https://hpc.nih.gov/storage/permissions.html | |||
=== No Automatic Backups === | |||
{{ClusterDataBackup}} | |||
=== Home directories === | |||
Your home directory on Cheaha is NFS-mounted to the compute nodes as /home/$USER or $HOME. It is acceptable to use your home directory as a location to store job scripts and custom code. You are responsible for keeping your home directory under 10GB in size! | |||
'''The home directory must not be used to store large amounts of data.''' Please use $USER_SCRATCH | |||
for actively used data sets and $USER_DATA for storage of non scratch data. | |||
=== Scratch === | |||
Research Computing policy requires that all bulky input and output must be located on the scratch space. The home directory is intended to store your job scripts, log files, libraries and other supporting files. | |||
'''Important Information:''' | |||
* Scratch space (network and local) '''is not backed up'''. | |||
* Research Computing expects each user to keep their scratch areas clean. The cluster scratch area are not to be used for archiving data. | |||
Cheaha has two types of scratch space, network mounted and local. | |||
* Network scratch ($USER_SCRATCH) is available on the login node and each compute node. This storage is a GPFS high performance file system providing roughly 4.7PB of usable storage. This should be your jobs primary working directory, unless the job would benefit from local scratch (see below). | |||
* Local scratch is physically located on each compute node and is not accessible to the other nodes (including the login node). This space is useful if the job performs a lot of file I/O. Most of the jobs that run on our clusters do not fall into this category. Because the local scratch is inaccessible outside the job, it is important to note that you must move any data between local scratch to your network accessible scratch within your job. For example, step 1 in the job could be to copy the input from $USER_SCRATCH to ${USER_SCRATCH}, step 2 code execution, step 3 move the data back to $USER_SCRATCH. | |||
==== Network Scratch ==== | |||
Network scratch is available using the environment variable $USER_SCRATCH or directly by /data/scratch/$USER | |||
It is advisable to use the environment variable whenever possible rather than the hard coded path. | |||
==== Local Scratch ==== | |||
Each compute node has a local scratch directory that is accessible via the variable '''$LOCAL_SCRATCH'''. If your job performs a lot of file I/O, the job should use $LOCAL_SCRATCH rather than $USER_SCRATCH to prevent bogging down the network scratch file system. The amount of scratch space available is approximately 800GB. | |||
The $LOCAL_SCRATCH is a special temporary directory and it's important to note that this directory is deleted when the job completes, so the job script has to move the results to $USER_SCRATCH or other location prior to the job exiting. | |||
Note that $LOCAL_SCRATCH is only useful for jobs in which all processes run on the same compute node, so MPI jobs are not candidates for this solution. | |||
The following is an array job example that uses $LOCAL_SCRATCH by transferring the inputs into $LOCAL_SCRATCH at the beginning of the script and the result out of $LOCAL_SCRATCH at the end of the script. | |||
<pre> | |||
#!/bin/bash | |||
#SBATCH --array=1-10 | |||
#SBATCH --share | |||
#SBATCH --partition=express | |||
# | |||
# Name your job to make it easier for you to track | |||
# | |||
#SBATCH --job-name=R_array_job | |||
# | |||
# Set your error and output files | |||
# | |||
#SBATCH --error=R_array_job.err | |||
#SBATCH --output=R_array_job.out | |||
#SBATCH --ntasks=1 | |||
# | |||
# Tell the scheduler only need 10 minutes and the appropriate partition | |||
# | |||
#SBATCH --time=00:10:00 | |||
#SBATCH --mem-per-cpu=256 | |||
# | |||
# Set your email address and request notification when you job is complete or if it fails | |||
# | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=YOUR_EMAIL_ADDRESS | |||
module load R/3.2.0-goolf-1.7.20 | |||
echo "TMPDIR: $LOCAL_SCRATCH" | |||
cd $LOCAL_SCRATCH | |||
# Create a working directory under the special scheduler local scratch directory | |||
# using the array job's taskID | |||
mdkir $SLURM_ARRAY_TASK_ID | |||
cd $SLURM_ARRAY_TASK_ID | |||
# Next copy the input data to the local scratch | |||
echo "Copying input data from network scratch to $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID - $(date) | |||
# The input data in this case has a numerical file extension that | |||
# matches $SLURM_ARRAY_TASK_ID | |||
cp -a $USER_SCRATCH/GeneData/INP*.$SLURM_ARRAY_TASK_ID ./ | |||
echo "copied input data from network scratch to $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID - $(date) | |||
someapp -S 1 -D 10 -i INP*.$SLURM_ARRAY_TASK_ID -o geneapp.out.$SLURM_ARRAY_TASK_ID | |||
# Lastly copy the results back to network scratch | |||
echo "Copying results from local $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID to network - $(date) | |||
cp -a geneapp.out.$SLURM_ARRAY_TASK_ID $USER_SCRATCH/GeneData/ | |||
echo "Copied results from local $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID to network - $(date) | |||
</pre> | |||
=== Project Storage === | |||
Cheaha has a location where shared data can be stored called $SHARE_PROJECT. As with user scratch, this area '''is not backed up'''! | |||
This is helpful if a team of researchers must access the same data. Please open a help desk ticket to request a project directory under $SHARE_PROJECT. | |||
=== Uploading Data === | |||
{{SensitiveInformation}} | |||
Data can be moved onto the cluster (pushed) from a remote client (ie. you desktop) via SCP or SFTP. Data can also be downloaded to the cluster (pulled) by issuing transfer commands once you are logged into the cluster. Common transfer methods are `wget <URL>`, FTP, or SCP, and depend on how the data is made available from the data provider. | |||
Large data sets should be staged directly to your $USER_SCRATCH directory so as not to fill up $HOME. If you are working on a data set shared with multiple users, it's preferable to request space in $SHARE_PROJECT rather than duplicating the data for each user. | |||
== Environment Modules == | |||
[http://modules.sourceforge.net/ Environment Modules] is installed on Cheaha and should be used when constructing your job scripts if an applicable module file exists. Using the module command you can easily configure your environment for specific software packages without having to know the specific environment variables and values to set. Modules allows you to dynamically configure your environment without having to logout / login for the changes to take affect. | |||
If you find that specific software does not have a module, please submit a [http://etlab.eng.uab.edu/ helpdesk ticket] to request the module. | |||
* Cheaha supports bash completion for the module command. For example, type 'module' and press the TAB key twice to see a list of options: | |||
<pre> | |||
module TAB TAB | |||
add display initlist keyword refresh switch use | |||
apropos help initprepend list rm unload whatis | |||
avail initadd initrm load show unuse | |||
clear initclear initswitch purge swap update | |||
</pre> | |||
* To see the list of available modulefiles on the cluster, run the '''module avail''' command (note the example list below may not be complete!) or '''module load ''' followed by two tab key presses: | |||
<pre> | |||
module avail | |||
----------------------------------------------------------------------------------------- /cm/shared/modulefiles ----------------------------------------------------------------------------------------- | |||
acml/gcc/64/5.3.1 acml/open64-int64/mp/fma4/5.3.1 fftw2/openmpi/gcc/64/float/2.1.5 intel-cluster-runtime/ia32/3.8 netcdf/gcc/64/4.3.3.1 | |||
acml/gcc/fma4/5.3.1 blacs/openmpi/gcc/64/1.1patch03 fftw2/openmpi/open64/64/double/2.1.5 intel-cluster-runtime/intel64/3.8 netcdf/open64/64/4.3.3.1 | |||
acml/gcc/mp/64/5.3.1 blacs/openmpi/open64/64/1.1patch03 fftw2/openmpi/open64/64/float/2.1.5 intel-cluster-runtime/mic/3.8 netperf/2.7.0 | |||
acml/gcc/mp/fma4/5.3.1 blas/gcc/64/3.6.0 fftw3/openmpi/gcc/64/3.3.4 intel-tbb-oss/ia32/44_20160526oss open64/4.5.2.1 | |||
acml/gcc-int64/64/5.3.1 blas/open64/64/3.6.0 fftw3/openmpi/open64/64/3.3.4 intel-tbb-oss/intel64/44_20160526oss openblas/dynamic/0.2.15 | |||
acml/gcc-int64/fma4/5.3.1 bonnie++/1.97.1 gdb/7.9 iozone/3_434 openmpi/gcc/64/1.10.1 | |||
acml/gcc-int64/mp/64/5.3.1 cmgui/7.2 globalarrays/openmpi/gcc/64/5.4 lapack/gcc/64/3.6.0 openmpi/open64/64/1.10.1 | |||
acml/gcc-int64/mp/fma4/5.3.1 cuda75/blas/7.5.18 globalarrays/openmpi/open64/64/5.4 lapack/open64/64/3.6.0 pbspro/13.0.2.153173 | |||
acml/open64/64/5.3.1 cuda75/fft/7.5.18 hdf5/1.6.10 mpich/ge/gcc/64/3.2 puppet/3.8.4 | |||
acml/open64/fma4/5.3.1 cuda75/gdk/352.79 hdf5_18/1.8.16 mpich/ge/open64/64/3.2 rc-base | |||
acml/open64/mp/64/5.3.1 cuda75/nsight/7.5.18 hpl/2.1 mpiexec/0.84_432 scalapack/mvapich2/gcc/64/2.0.2 | |||
acml/open64/mp/fma4/5.3.1 cuda75/profiler/7.5.18 hwloc/1.10.1 mvapich/gcc/64/1.2rc1 scalapack/openmpi/gcc/64/2.0.2 | |||
acml/open64-int64/64/5.3.1 cuda75/toolkit/7.5.18 intel/compiler/32/15.0/2015.5.223 mvapich/open64/64/1.2rc1 sge/2011.11p1 | |||
acml/open64-int64/fma4/5.3.1 default-environment intel/compiler/64/15.0/2015.5.223 mvapich2/gcc/64/2.2b slurm/15.08.6 | |||
acml/open64-int64/mp/64/5.3.1 fftw2/openmpi/gcc/64/double/2.1.5 intel-cluster-checker/2.2.2 mvapich2/open64/64/2.2b torque/6.0.0.1 | |||
---------------------------------------------------------------------------------------- /share/apps/modulefiles ----------------------------------------------------------------------------------------- | |||
rc/BrainSuite/15b rc/freesurfer/freesurfer-5.3.0 rc/intel/compiler/64/ps_2016/2016.0.047 rc/matlab/R2015a rc/SAS/v9.4 | |||
rc/cmg/2012.116.G rc/gromacs-intel/5.1.1 rc/Mathematica/10.3 rc/matlab/R2015b | |||
rc/dsistudio/dsistudio-20151020 rc/gtool/0.7.5 rc/matlab/R2012a rc/MRIConvert/2.0.8 | |||
--------------------------------------------------------------------------------------- /share/apps/rc/modules/all --------------------------------------------------------------------------------------- | |||
AFNI/linux_openmp_64-goolf-1.7.20-20160616 gperf/3.0.4-intel-2016a MVAPICH2/2.2b-GCC-4.9.3-2.25 | |||
Amber/14-intel-2016a-AmberTools-15-patchlevel-13-13 grep/2.15-goolf-1.4.10 NASM/2.11.06-goolf-1.7.20 | |||
annovar/2016Feb01-foss-2015b-Perl-5.22.1 GROMACS/5.0.5-intel-2015b-hybrid NASM/2.11.08-foss-2015b | |||
ant/1.9.6-Java-1.7.0_80 GSL/1.16-goolf-1.7.20 NASM/2.11.08-intel-2016a | |||
APBS/1.4-linux-static-x86_64 GSL/1.16-intel-2015b NASM/2.12.02-foss-2016a | |||
ASHS/rev103_20140612 GSL/2.1-foss-2015b NASM/2.12.02-intel-2015b | |||
Aspera-Connect/3.6.1 gtool/0.7.5_linux_x86_64 NASM/2.12.02-intel-2016a | |||
ATLAS/3.10.1-gompi-1.5.12-LAPACK-3.4.2 guile/1.8.8-GNU-4.9.3-2.25 ncurses/5.9-foss-2015b | |||
Autoconf/2.69-foss-2016a HAPGEN2/2.2.0 ncurses/5.9-GCC-4.8.4 | |||
Autoconf/2.69-GCC-4.8.4 HarfBuzz/1.2.7-intel-2016a ncurses/5.9-GNU-4.9.3-2.25 | |||
Autoconf/2.69-GNU-4.9.3-2.25 HDF5/1.8.15-patch1-intel-2015b ncurses/5.9-goolf-1.4.10 | |||
. | |||
. | |||
. | |||
. | |||
</pre> | |||
Some software packages have multiple module files, for example: | |||
* GCC/4.7.2 | |||
* GCC/4.8.1 | |||
* GCC/4.8.2 | |||
* GCC/4.8.4 | |||
* GCC/4.9.2 | |||
* GCC/4.9.3 | |||
* GCC/4.9.3-2.25 | |||
In this case, the GCC module will always load the latest version, so loading this module is equivalent to loading GCC/4.9.3-2.25. If you always want to use the latest version, use this approach. If you want use a specific version, use the module file containing the appropriate version number. | |||
Some modules, when loaded, will actually load other modules. For example, the ''GROMACS/5.0.5-intel-2015b-hybrid '' module will also load ''intel/2015b'' and other related tools. | |||
* To load a module, ex: for a GROMACS job, use the following '''module load''' command in your job script: | |||
<pre> | |||
module load GROMACS/5.0.5-intel-2015b-hybrid | |||
</pre> | |||
* To see a list of the modules that you currently have loaded use the '''module list''' command | |||
<pre> | |||
module list | |||
Currently Loaded Modulefiles: | |||
1) slurm/15.08.6 9) impi/5.0.3.048-iccifort-2015.3.187-GNU-4.9.3-2.25 17) Tcl/8.6.3-intel-2015b | |||
2) rc-base 10) iimpi/7.3.5-GNU-4.9.3-2.25 18) SQLite/3.8.8.1-intel-2015b | |||
3) GCC/4.9.3-binutils-2.25 11) imkl/11.2.3.187-iimpi-7.3.5-GNU-4.9.3-2.25 19) Tk/8.6.3-intel-2015b-no-X11 | |||
4) binutils/2.25-GCC-4.9.3-binutils-2.25 12) intel/2015b 20) Python/2.7.9-intel-2015b | |||
5) GNU/4.9.3-2.25 13) bzip2/1.0.6-intel-2015b 21) Boost/1.58.0-intel-2015b-Python-2.7.9 | |||
6) icc/2015.3.187-GNU-4.9.3-2.25 14) zlib/1.2.8-intel-2015b 22) GROMACS/5.0.5-intel-2015b-hybrid | |||
7) ifort/2015.3.187-GNU-4.9.3-2.25 15) ncurses/5.9-intel-2015b | |||
8) iccifort/2015.3.187-GNU-4.9.3-2.25 16) libreadline/6.3-intel-2015b | |||
</pre> | |||
* A module can be removed from your environment by using the '''module unload''' command: | |||
<pre> | |||
module unload GROMACS/5.0.5-intel-2015b-hybrid | |||
</pre> | |||
* The definition of a module can also be viewed using the '''module show''' command, revealing what a specific module will do to your environment: | |||
<pre> | |||
module show GROMACS/5.0.5-intel-2015b-hybrid | |||
------------------------------------------------------------------- | |||
/share/apps/rc/modules/all/GROMACS/5.0.5-intel-2015b-hybrid: | |||
module-whatis GROMACS is a versatile package to perform molecular dynamics, | |||
i.e. simulate the Newtonian equations of motion for systems with hundreds to millions of particles. - Homepage: http://www.gromacs.org | |||
conflict GROMACS | |||
prepend-path CPATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/include | |||
prepend-path LD_LIBRARY_PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/lib64 | |||
prepend-path LIBRARY_PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/lib64 | |||
prepend-path MANPATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/share/man | |||
prepend-path PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/bin | |||
prepend-path PKG_CONFIG_PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/lib64/pkgconfig | |||
setenv EBROOTGROMACS /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid | |||
setenv EBVERSIONGROMACS 5.0.5 | |||
setenv EBDEVELGROMACS /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/easybuild/GROMACS-5.0.5-intel-2015b-hybrid-easybuild-devel | |||
------------------------------------------------------------------- | |||
</pre> | |||
=== Error Using Modules from a Job Script === | |||
If you are using modules and the command your job executes runs fine from the command line but fails when you run it from the job, you may be having an issue with the script initialization. If you see this error in your job error output file | |||
<pre> | |||
-bash: module: line 1: syntax error: unexpected end of file | |||
-bash: error importing function definition for `BASH_FUNC_module' | |||
</pre> | |||
Add the command `unset module` before calling your module files. The -V job argument will cause a conflict with the module function used in your script. | |||
== Sample Job Scripts == | |||
The following are sample job scripts, please be careful to edit these for your environment (i.e. replace <font color="red">YOUR_EMAIL_ADDRESS</font> with your real email address), set the h_rt to an appropriate runtime limit and modify the job name and any other parameters. | |||
'''Hello World''' is the classic example used throughout programming. We don't want to buck the system, so we'll use it as well to demonstrate jobs submission with one minor variation: our hello world will send us a greeting using the name of whatever machine it runs on. For example, when run on the Cheaha login node, it would print "Hello from login001". | |||
=== Hello World (serial) === | |||
A serial job is one that can run independently of other commands, ie. it doesn't depend on the data from other jobs running simultaneously. You can run many serial jobs in any order. This is a common solution to processing lots of data when each command works on a single piece of data. For example, running the same conversion on 100s of images. | |||
Here we show how to create job script for one simple command. Running more than one command just requires submitting more jobs. | |||
* Create your hello world application. Run this command to create a script, turn it into to a command, and run the command (just copy and past the following on to the command line). | |||
1. Create the file: | |||
<pre> | |||
$ vim helloworld.sh | |||
</pre> | |||
2. Write into "helloworld.sh" file (To write in vim editor: press '''shift + I''' ) | |||
<pre> | |||
#!/bin/bash | |||
echo Hello from `hostname` | |||
</pre> | |||
3. Save the file by pressing the '''esc''' key, type the following | |||
<pre> | |||
:wq | |||
</pre> | |||
4. Need to give permission the "helloworld.sh" file | |||
<pre> | |||
$ chmod +x helloworld.sh | |||
</pre> | |||
* Create the Slurm job script that will request 256 MB RAM and a maximum runtime of 10 minutes. | |||
1. Create the JOB file: | |||
<pre> | |||
$ vim helloworld.job | |||
</pre> | |||
2. Write into "helloworld.job" file (To write in vim editor: press '''shift + I''' ) | |||
<pre> | |||
#!/bin/bash | |||
#SBATCH --share | |||
#SBATCH --partition=express | |||
# | |||
# Name your job to make it easier for you to track | |||
# | |||
#SBATCH --job-name=helloworld | |||
# | |||
# Set your error and output files | |||
# | |||
#SBATCH --error=helloworld.err | |||
#SBATCH --output=helloworld.out | |||
#SBATCH --ntasks=1 | |||
# | |||
# Tell the scheduler only need 10 minutes | |||
# | |||
#SBATCH --time=00:10:00 | |||
#SBATCH --mem-per-cpu=256 | |||
# | |||
# Set your email address and request notification when you job is complete or if it fails | |||
# | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=$USER@uab.edu | |||
./helloworld.sh | |||
</pre> | |||
3. Save the file by pressing the '''esc''' key, type the following | |||
<pre> | |||
:wq | |||
</pre> | |||
* Submit the job to Slurm scheduler and check the status using squeue | |||
<pre> | |||
$ sbatch helloworld.job | |||
Submitted batch job 52888 | |||
</pre> | |||
* When the job completes, you should have output files named helloworld.out and helloworld.err | |||
<pre> | |||
$ cat helloworld.out | |||
Hello from c0003 | |||
</pre> | |||
=== Hello World (parallel with MPI) === | |||
MPI is used to coordinate the activity of many computations occurring in parallel. It is commonly used in simulation software for molecular dynamics, fluid dynamics, and similar domains where there is significant communication (data) exchanged between cooperating process. | |||
Here is a simple parallel Slurm job script for running commands the rely on MPI. This example also includes the example of compiling the code and submitting the job script to the Slurm scheduler. | |||
* First, create a directory for the Hello World jobs | |||
<pre> | |||
$ mkdir -p ~/jobs/helloworld | |||
$ cd ~/jobs/helloworld | |||
</pre> | |||
* Create the Hello World code written in C (this example of MPI enabled Hello World includes a 3 minute sleep to ensure the job runs for several minutes, a normal hello world example would run in a matter of seconds). | |||
<pre> | |||
$ vi helloworld-mpi.c | |||
</pre> | |||
<pre> | |||
#include <stdio.h> | |||
#include <mpi.h> | |||
main(int argc, char **argv) | |||
{ | |||
int rank, size; | |||
int i, j; | |||
float f; | |||
MPI_Init(&argc,&argv); | |||
MPI_Comm_rank(MPI_COMM_WORLD, &rank); | |||
MPI_Comm_size(MPI_COMM_WORLD, &size); | |||
printf("Hello World from process %d of %d.\n", rank, size); | |||
sleep(180); | |||
for (j=0; j<=100000; j++) | |||
for(i=0; i<=100000; i++) | |||
f=i*2.718281828*i+i+i*3.141592654; | |||
MPI_Finalize(); | |||
} | |||
</pre> | |||
* Compile the code, first purging any modules you may have loaded followed by loading the module for OpenMPI GNU. The mpicc command will compile the code and produce a binary named helloworld_gnu_openmpi | |||
<pre> | |||
$ module purge | |||
$ module load DefaultModules | |||
$ module load OpenMPI/4.0.1-GCC-8.3.0-2.32 | |||
$ mpicc helloworld-mpi.c -o helloworld_gnu_openmpi | |||
</pre> | |||
* Create the Slurm job script that will request 8 cpu slots and a maximum runtime of 10 minutes | |||
<pre> | |||
$ vi helloworld.job | |||
</pre> | |||
<pre> | |||
#!/bin/bash | |||
#SBATCH --share | |||
#SBATCH --partition=express | |||
# | |||
# Name your job to make it easier for you to track | |||
# | |||
#SBATCH --job-name=helloworld_mpi | |||
# | |||
# Set your error and output files | |||
# | |||
#SBATCH --error=helloworld_mpi.err | |||
#SBATCH --output=helloworld_mpi.out | |||
#SBATCH --ntasks=8 | |||
# | |||
# Tell the scheduler only need 10 minutes | |||
# | |||
#SBATCH --time=00:10:00 | |||
#SBATCH --mem-per-cpu=256 | |||
# | |||
# Set your email address and request notification when you job is complete or if it fails | |||
# | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=YOUR_EMAIL_ADDRESS | |||
module load OpenMPI/1.8.8-GNU-4.9.3-2.25 | |||
mpirun -np $SLURM_NTASKS helloworld_gnu_openmpi | |||
</pre> | |||
* Submit the job to Slurm scheduler and check the status using squeue -u $USER | |||
<pre> | |||
$ sbatch helloworld.job | |||
Submitted batch job 52893 | |||
$ squeue -u BLAZERID | |||
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON) | |||
52893 express hellowor BLAZERID R 2:07 2 c[0005-0006] | |||
</pre> | |||
* When the job completes, you should have output files named helloworld_mpi.out and helloworld_mpi.err | |||
<pre> | |||
$ cat helloworld_mpi.out | |||
Hello World from process 1 of 8. | |||
Hello World from process 3 of 8. | |||
Hello World from process 4 of 8. | |||
Hello World from process 7 of 8. | |||
Hello World from process 5 of 8. | |||
Hello World from process 6 of 8. | |||
Hello World from process 0 of 8. | |||
Hello World from process 2 of 8. | |||
</pre> | |||
=== Hello World (serial) -- revisited === | |||
The job submit scripts (sbatch scripts) are actually bash shell scripts in their own right. The reason for using the funky #SBATCH prefix in the scripts is so that bash interprets any such line as a comment and won't execute it. Because the # character starts a comment in bash, we can weave the Slurm scheduler directives (the #SBATCH lines) into standard bash scripts. This lets us build scripts that we can execute locally and then easily run the same script to on a cluster node by calling it with sbatch. This can be used to our advantage to create a more fluid experience in moving between development and production job runs. | |||
The following example is a simple variation on the serial job above. All we will do is convert our Slurm job script into a command called helloworld that calls the helloworld.sh command. | |||
If the first line of a file is #!/bin/bash and that file is executable, the shell will automatically run the command as if were any other system command, eg. ls. That is, the ".sh" extension on our HelloWorld.sh script is completely optional and is only meaningful to the user. | |||
Copy the serial helloworld.job script to a new file, add a the special #!/bin/bash as the first line, and make it executable with the following command (note: those are single quotes in the echo command): | |||
<pre> | |||
echo '#!/bin/bash' | cat helloworld.job > helloworld ; chmod +x helloworld | |||
</pre> | |||
Our sbatch script has now become a regular command. We can now execute the command with the simple prefix "./helloworld", which means "execute this file in the current directory": | |||
<pre> | |||
./helloworld | |||
Hello from login001 | |||
</pre> | |||
Or if we want to run the command on a compute node, replace the "./" prefix with "sbatch ": | |||
<pre> | |||
$ sbatch helloworld | |||
Submitted batch job 53001 | |||
</pre> | |||
And when the cluster run is complete you can look at the content of the output: | |||
<pre> | |||
$ $ cat helloworld.out | |||
Hello from c0003 | |||
</pre> | |||
You can use this approach of treating you sbatch files as command wrappers to build a collection of commands that can be executed locally or via sbatch. The other examples can be restructured similarly. | |||
To avoid having to use the "./" prefix, just add the current directory to your PATH. Also, if you plan to do heavy development using this feature on the cluster, please be sure to run [https://docs.uabgrid.uab.edu/wiki/Slurm#Interactive_Session sinteractive] first so you don't load the login node with your development work. | |||
=== Gromacs === | |||
<pre> | |||
#!/bin/bash | |||
#SBATCH --partition=short | |||
# | |||
# Name your job to make it easier for you to track | |||
# | |||
#SBATCH --job-name=test_gromacs | |||
# | |||
# Set your error and output files | |||
# | |||
#SBATCH --error=test_gromacs.err | |||
#SBATCH --output=test_gromacs.out | |||
#SBATCH --ntasks=8 | |||
# | |||
# Tell the scheduler only need 10 minutes | |||
# | |||
#SBATCH --time=10:00:00 | |||
#SBATCH --mem-per-cpu=2048 | |||
# | |||
# Set your email address and request notification when you job is complete or if it fails | |||
# | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=YOUR_EMAIL_ADDRESS | |||
module load OpenMPI/1.8.8-GNU-4.9.3-2.25 | |||
module load GROMACS/5.0.5-intel-2015b-hybrid | |||
# Change directory to the job working directory if not already there | |||
cd ${USER_SCRATCH}/jobs/gromacs | |||
# Single precision | |||
MDRUN=mdrun_mpi | |||
# Enter your tpr file over here | |||
export MYFILE=example.tpr | |||
mpirun -np SLURM_NTASKS $MDRUN -v -s $MYFILE -o $MYFILE -c $MYFILE -x $MYFILE -e $MYFILE -g ${MYFILE}.log | |||
</pre> | |||
=== R (array job) === | |||
The following is an example job script that will use an array of 10 tasks (--array=1-10), each task has a max runtime of 2 hours and will use no more than 256 MB of RAM per task. Array's of tasks are useful when you have lots of simple jobs that work on their own separate files or a sub-set of the problem that can be selected by the array task index. For [https://gitlab.rc.uab.edu/rc-training-sessions/Tutorial_parallelism a more comprehensive introduction please see this tutorial]. | |||
Create a working directory and the job submission script | |||
<pre> | |||
$ mkdir -p ~/jobs/ArrayExample | |||
$ cd ~/jobs/ArrayExample | |||
$ vi R-example-array.job | |||
</pre> | |||
<pre> | |||
#!/bin/bash | |||
#SBATCH --array=1-10 | |||
#SBATCH --share | |||
#SBATCH --partition=express | |||
# | |||
# Name your job to make it easier for you to track | |||
# | |||
#SBATCH --job-name=R_array_job | |||
# | |||
# Set your error and output files | |||
# | |||
#SBATCH --error=R_array_job.err | |||
#SBATCH --output=R_array_job.out | |||
#SBATCH --ntasks=1 | |||
# | |||
# Tell the scheduler only need 10 minutes | |||
# | |||
#SBATCH --time=00:10:00 | |||
#SBATCH --mem-per-cpu=256 | |||
# | |||
# Set your email address and request notification when you job is complete or if it fails | |||
# | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=YOUR_EMAIL_ADDRESS | |||
module load R/3.2.0-goolf-1.7.20 | |||
cd ~/jobs/ArrayExample/rep$SLURM_ARRAY_TASK_ID | |||
srun R CMD BATCH rscript.R | |||
</pre> | |||
Submit the job to the Slurm scheduler and check the status of the job using the squeue command | |||
<pre> | |||
$ sbatch R-example-array.job | |||
$ squeue -u $USER | |||
</pre> | |||
=== Array Job Parameterization === | |||
Suppose you need to submit thousands of jobs. While you could do this in a for loop, the global limit on jobs in the SLURM queue is 10,000. The limit is in place for performance reasons and the jobs may be rejected with the following error message and an incomplete set of tasks. | |||
<pre>sbatch: error: Slurm temporarily unable to accept job, sleeping and retrying</pre> | |||
The preferred way to handle this scenario is to allow SLURM to schedule the jobs for you using the array flag in an sbatch script. This allows many jobs to be submitted as a single entry in the queue, letting SLURM handle the for loop and queueing. It is possible to reference the current loop index, or task id, as $SLURM_ARRAY_TASK_ID. | |||
An example using $SLURM_ARRAY_TASK_ID to load input files and create output files is shown below. Suppose you have a short script called my_processing_script that needs to be run on 20,000 separate files. Suppose each instance only needs 1 cpu and 2 GB of RAM and finishes in 5 minutes. Submitting these files all at once won't work and at least half of them will be rejected by SLURM. Instead we can use the sbatch array flag. Note that some other useful flags have been omitted for brevity. | |||
<pre> | |||
#! /bin/bash | |||
#SBATCH --partition=express | |||
#SBATCH --ntasks=1 | |||
#SBATCH --cpus-per-task=1 | |||
#SBATCH --mem-per-cpu=2G | |||
#SBATCH --array=1-20000%100 | |||
# This will run tasks 1 through 20000, with up to 100 at a time. | |||
# It is possible to provide any comma-separated list of intervals. | |||
# An example of a valid subset is --array=1,2,5-1000,3777,4995-5000%100 | |||
INPUT_FILE=$USER_DATA/input/file_$SLURM_ARRAY_TASK_ID.txt | |||
OUTPUT_FILE=$USER_DATA/output/file_$SLURM_ARRAY_TASK_ID.txt | |||
my_processing_script --input="$INPUT_FILE" --output="$OUTPUT_FILE" | |||
</pre> | |||
=== GPU JOB === | |||
A Graphics processing unit (GPU) is a specialized electronic circuit designed to rapidly manipulate and alter memory to accelerate the creation of images in a frame buffer intended for output to a display device. | |||
Create a math.sh file as: | |||
<pre> | |||
$vim math.sh | |||
</pre> | |||
<pre> | |||
#!/bin/bash | |||
(e=5) | |||
echo $e | |||
(( e = e + 3 )) | |||
echo $e | |||
(( e=e+4 )) # -- spaces or no spaces, it doesn't matter | |||
echo $e | |||
</pre> | |||
Give File permissions for script as follows: | |||
<pre> | |||
$chmod +x math.sh | |||
</pre> | |||
Create Job submission script file: | |||
<pre> | |||
$vi math.job | |||
</pre> | |||
<pre> | |||
#!/bin/bash | |||
#SBATCH --share | |||
#SBATCH --partition=pascalnodes | |||
#SBATCH --gres=gpu:1 | |||
# Name your job to make it easier for you to track | |||
# | |||
#SBATCH --job-name=math | |||
# | |||
# Set your error and output files | |||
# | |||
#SBATCH --error=math.err | |||
#SBATCH --output=math.out | |||
#SBATCH --ntasks=1 | |||
# | |||
# Tell the scheduler only need 10 minutes | |||
# | |||
#SBATCH --time=00:10:00 | |||
#SBATCH --mem-per-cpu=256 | |||
# | |||
# Set your email address and request notification when you job is complete or if it fails | |||
# | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=$USER@uab.edu | |||
./math.sh | |||
</pre> | |||
Submitting batch script to Slurm scheduler | |||
<pre> | |||
$sbatch math.job | |||
</pre> | |||
We can also request GPU's on cluster as: | |||
<pre> | |||
$sinteractive --ntasks=1 --time=00:10:00 --exclusive --partition=pascalnodes -N2 --gres=gpu:2 | |||
</pre> | |||
=== GPU Job (with MPI) === | |||
As mentioned above, MPI is used to coordinate the activity of many computations occurring in parallel. It is commonly used in simulation software for molecular dynamics, fluid dynamics, and similar domains where there is significant communication (data) exchanged between cooperating process. | |||
An example of an GPU job with MPI can be found by visiting [https://gitlab.rc.uab.edu/wsmonroe/horovod-environment/blob/master/README.md this link]. | |||
Be sure to request the appropiate amount of gpu resources for your job: | |||
<pre> | |||
sinteractive --ntasks=8 --time=08:00:00 --exclusive --partition=pascalnodes -N2 --gres=gpu:4 | |||
</pre> | |||
=== Singularity Container === | |||
Singularity is designed so that you can use it within SLURM jobs and it does not violate security constraints on the cluster. Singularity was built keeping HPC in mind, i.e a shared environment. Using Singularity container with SLURM job script is very easy, as the containers run as a process on the host machine, just like any other command in a batch script. You just need to load Singularity in your job script and run the command via a singularity process. Here's an example job script below: | |||
<pre> | |||
#!/bin/bash | |||
# | |||
#SBATCH --job-name=test-singularity | |||
#SBATCH --output=res.out | |||
#SBATCH --error=res.err | |||
# | |||
# Number of tasks needed for this job. Generally, used with MPI jobs | |||
#SBATCH --ntasks=1 | |||
#SBATCH --partition=express | |||
# | |||
# Time format = HH:MM:SS, DD-HH:MM:SS | |||
#SBATCH --time=10:00 | |||
# | |||
# Number of CPUs allocated to each task. | |||
#SBATCH --cpus-per-task=1 | |||
# | |||
# Mimimum memory required per allocated CPU in MegaBytes. | |||
#SBATCH --mem-per-cpu=100 | |||
# | |||
# Send mail to the email address when the job fails | |||
#SBATCH --mail-type=FAIL | |||
#SBATCH --mail-user=$USER@uab.edu | |||
#Set your environment here | |||
module load Singularity/2.5.2-GCC-5.4.0-2.26 | |||
#Run your singularity or any other commands here | |||
singularity exec -B /data/user/$USER /data/user/$USER/rc-training-sessions/neurodebian-neurodebian-master-latest.simg dcm2nii PATH_TO_YOUR_DICOM_FILES | |||
</pre> | |||
For [https://gitlab.rc.uab.edu/rc-training-sessions/singularity_containers a more comprehensive introduction please see this tutorial]. | |||
== Installed Software == | |||
A partial list of installed software with additional instructions for their use is available on the [[Cheaha Software]] page. | |||
Latest revision as of 17:11, 30 August 2022
Cheaha is a cluster computing environment for UAB researchers. Information about the history and future plans for Cheaha is available on the Cheaha page.
Access (Cluster Account Request)
To request an account on Cheaha, please send an email to support@listserv.uab.edu to request an account. Please include some background information about the work you plan on doing on the cluster and the group you work with, ie. your lab or affiliation.
NOTE: The email you send to support@listserv.uab.edu will send back a confirmation email which must be acknowledged in order to submit the account request. This additional step is meant to cut down on spam to the support list and is only needed for the initial account creation request sent to the support list.
Usage of Cheaha is governed by UAB's Acceptable Use Policy (AUP) for computer resources.
External Collaborator
To request an account for an external collaborator, please follow the steps here.
Login
Overview
Once your account has been created, you'll receive an email containing your user ID, generally your Blazer ID. You can log into Cheaha via your web browser using the new web-based HPC experience.
You can also log into Cheaha via a traditional SSH client. Most UAB Windows workstations already have an SSH client installed, possibly named SSH Secure Shell Client or PuTTY. Linux and Mac OS X systems should have an SSH client installed by default.
Usage of Cheaha is governed by UAB's Acceptable Use Policy (AUP) for computer and network resources.
Client Configuration
This section will cover steps to configure Windows, Linux and Mac OS X clients to connect to Cheaha.
The official DNS name of Cheaha's frontend machine is cheaha.rc.uab.edu. If you want to refer to the machine as cheaha, you'll have to either add the "rc.uab.edu" to you computer's DNS search path. On Unix-derived systems (Linux, Mac) you can edit your computers /etc/resolv.conf as follows (you'll need administrator access to edit this file)
search rc.uab.edu
Or you can customize your SSH configuration to use the short name "cheaha" as a connection name. On systems using OpenSSH you can add the following to your ~/.ssh/config file
Host cheaha Hostname cheaha.rc.uab.edu
Linux
Linux systems, regardless of the flavor (RedHat, SuSE, Ubuntu, etc...), should already have an SSH client on the system as part of the default install.
- Start a terminal (on RedHat click Applications -> Accessories -> Terminal, on Ubuntu Ctrl+Alt+T)
- At the prompt, enter the following command to connect to Cheaha (Replace blazerid with your Cheaha userid)
ssh blazerid@cheaha.rc.uab.edu
Mac OS X
Mac OS X is a Unix operating system (BSD) and has a built in ssh client.
- Start a terminal (click Finder, type Terminal and double click on Terminal under the Applications category)
- At the prompt, enter the following command to connect to Cheaha (Replace blazerid with your Cheaha userid)
ssh blazerid@cheaha.rc.uab.edu
Windows
There are many SSH clients available for Windows, some commercial and some that are free (GPL). This section will cover two clients that are commonly found on UAB Windows systems.
MobaXterm
MobaXterm is a free (also available for a price in a Profession version) suite of SSH tools. Of the Windows clients we've used, MobaXterm is the easiest to use and feature complete. Features include (but not limited to):
- SSH client (in a handy web browser like tabbed interface)
- Embedded Cygwin (which allows Windows users to run many Linux commands like grep, rsync, sed)
- Remote file system browser (graphical SFTP)
- X11 forwarding for remotely displaying graphical content from Cheaha
- Installs without requiring Windows Administrator rights
Start MobaXterm and click the Session toolbar button (top left). Click SSH for the session type, enter the following information and click OK. Once finished, double click cheaha.rc.uab.edu in the list of Saved sessions under PuTTY sessions:
| Field | Cheaha Settings |
|---|---|
| Remote host | cheaha.rc.uab.edu |
| Port | 22 |
PuTTY
PuTTY is a free suite of SSH and telnet tools written and maintained by Simon Tatham. PuTTY supports SSH, secure FTP (SFTP), and X forwarding (XTERM) among other tools.
- Start PuTTY (Click START -> All Programs -> PuTTY -> PuTTY). The 'PuTTY Configuration' window will open
- Use these settings for each of the clusters that you would like to configure
| Field | Cheaha Settings |
|---|---|
| Host Name (or IP address) | cheaha.rc.uab.edu |
| Port | 22 |
| Protocol | SSH |
| Saved Sessions | cheaha.rc.uab.edu |
- Click Save to save the configuration, repeat the previous steps for the other clusters
- The next time you start PuTTY, simply double click on the cluster name under the 'Saved Sessions' list
SSH Secure Shell Client
SSH Secure Shell is a commercial application that is installed on many Windows workstations on campus and can be configured as follows:
- Start the program (Click START -> All Programs -> SSH Secure Shell -> Secure Shell Client). The 'default - SSH Secure Shell' window will open
- Click File -> Profiles -> Add Profile to open the 'Add Profile' window
- Type in the name of the cluster (for example: cheaha) in the field and click 'Add to Profiles'
- Click File -> Profiles -> Edit Profiles to open the 'Profiles' window
- Single click on your new profile name
- Use these settings for the clusters
| Field | Cheaha Settings |
|---|---|
| Host name | cheaha.rc.uab.edu |
| User name | blazerid (insert your blazerid here) |
| Port | 22 |
| Protocol | SSH |
| Encryption algorithm | <Default> |
| MAC algorithm | <Default> |
| Compression | <None> |
| Terminal answerback | vt100 |
- Leave 'Connect through firewall' and 'Request tunnels only' unchecked
- Click OK to save the configuration, repeat the previous steps for the other clusters
- The next time you start SSH Secure Shell, click 'Profiles' and click the cluster name
Logging in to Cheaha
No matter which client you use to connect to the Cheaha, the first time you connect, the SSH client should display a message asking if you would like to import the hosts public key. Answer Yes to this question.
- Connect to Cheaha using one of the methods listed above
- Answer Yes to import the cluster's public key
- Enter your BlazerID password
- After successfully logging in for the first time, You may see the following message just press ENTER for the next three prompts, don't type any passphrases!
It doesn't appear that you have set up your ssh key.
This process will make the files:
/home/joeuser/.ssh/id_rsa.pub
/home/joeuser/.ssh/id_rsa
/home/joeuser/.ssh/authorized_keys
Generating public/private rsa key pair.
Enter file in which to save the key (/home/joeuser/.ssh/id_rsa):
- Enter file in which to save the key (/home/joeuser/.ssh/id_rsa):Press Enter
- Enter passphrase (empty for no passphrase):Press Enter
- Enter same passphrase again:Press Enter
Your identification has been saved in /home/joeuser/.ssh/id_rsa. Your public key has been saved in /home/joeuser/.ssh/id_rsa.pub. The key fingerprint is: f6:xx:xx:xx:xx:dd:9a:79:7b:83:xx:f9:d7:a7:d6:27 joeuser@cheaha.rc.uab.edu
Users without a blazerid (collaborators from other universities)
- If you were issued a temporary password, enter it (Passwords are CaSE SensitivE!!!) You should see a message similar to this
You are required to change your password immediately (password aged)
WARNING: Your password has expired. You must change your password now and login again! Changing password for user joeuser. Changing password for joeuser (current) UNIX password:
- (current) UNIX password: Enter your temporary password at this prompt and press enter
- New UNIX password: Enter your new strong password and press enter
- Retype new UNIX password: Enter your new strong password again and press enter
- After you enter your new password for the second time and press enter, the shell may exit automatically. If it doesn't, type exit and press enter
- Log in again, this time use your new password
Congratulations, you should now have a command prompt and be ready to start submitting jobs!!!
Hardware
The Cheaha Compute Platform includes commodity compute hardware, totaling 2800 compute cores and over 4.7PB of usable storage (6.6PB raw capacity). The following descriptions highlight the current hardware profile that provides an aggregate theoretical peak performance of 468 teraflops.
- Compute
- 36 Compute Nodes with two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 128GB DDR4 RAM, FDR InfiniBand and 10GigE network cards
- 38 Compute Nodes with two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 256GB DDR4 RAM, FDR InfiniBand and 10GigE network cards
- 14 Compute Nodes with two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 384GB DDR4 RAM, FDR InfiniBand and 10GigE network cards
- 4 Compute Nodes with Nvidia Tesla K80 and two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 128GB DDR4 RAM, FDR InfiniBand and 10GigE network cards
- 4 Compute Nodes with Intel Phi coprocessor SE10/7120 and two 12 core processors (Intel Xeon E5-2680 v3 2.5GHz) with 128GB DDR4 RAM, FDR InfiniBand and 10GigE network cards
- 18 Compute Nodes with two 14 core processors (Intel Xeon E5-2680 v4 2.4GHz)with 256GB DDR4 RAM, four NVIDIA Tesla P100 16GB GPUs, EDR InfiniBand and 10GigE network cards
- Networking
- FDR and EDR InfiniBand Switch
- 10Gigabit Ethernet Switch
- Storage -- DDN SFA12KX with GPFS)
- 2 x 12KX40D-56IB controllers
- 10 x SS8460 disk enclosures
- 825 x 4K SAS drives
- Management
- Management node and gigabit switch for cluster management
- Bright Advanced Cluster Management software licenses
Cluster Software
- BrightCM 7.2
- CentOS 7.2 x86_64
- Slurm 15.08
Queuing System
All work on Cheaha must be submitted to our queuing system (Slurm). A common mistake made by new users is to run 'jobs' on the login node. This section gives a basic overview of what a queuing system is and why we use it.
What is a queuing system?
- Software that gives users fair allocation of the cluster's resources
- Schedules jobs based using resource requests (the following are commonly requested resources, there are many more that are available)
- Number of processors (often referred to as "slots")
- Maximum memory (RAM) required per slot
- Maximum run time
- Common queuing systems:
- Slurm
- Sun Grid Engine (Also know as SGE, OGE, GE)
- OpenPBS
- Torque
- LSF (load sharing facility)
Slurm is a queue management system and stands for Simple Linux Utility for Resource Management. Slurm was developed at the Lawrence Livermore National Lab and currently runs some of the largest compute clusters in the world. Slurm is now the primary job manager on Cheaha, it replaces SUN Grid Engine ([SGE]) the job manager used earlier. Instructions of using SLURM and writing SLURM scripts for jobs submission on Cheaha can be found here.
Typical Workflow
- Stage data to $USER_SCRATCH (your scratch directory)
- Research how to run your code in "batch" mode. Batch mode typically means the ability to run it from the command line without requiring any interaction from the user.
- Identify the appropriate resources needed to run the job. The following are mandatory resource requests for all jobs on Cheaha
- Maximum memory (RAM) required per slot
- Maximum runtime
- Write a job script specifying queuing system parameters, resource requests and commands to run program
- Submit script to queuing system (sbatch script.job)
- Monitor job (squeue)
- Review the results and resubmit as necessary
- Clean up the scratch directory by moving or deleting the data off of the cluster
Resource Requests
Accurate resource requests are extremely important to the health of the over all cluster. In order for Cheaha to operate properly, the queing system must know how much runtime and RAM each job will need.
Mandatory Resource Requests
- -t, --time=
Set a limit on the total run time of the job allocation. If the requested time limit exceeds the partition's time limit, the job will be left in a PENDING state (possibly indefinitely).
- For Array jobs, this represents the maximum run time for each task
- For serial or parallel jobs, this represents the maximum run time for the entire job
- --mem-per-cpu=<MB>
Mimimum memory required per allocated CPU in MegaBytes.
Other Common Resource Requests
- -N, --nodes=<minnodes[-maxnodes]>
Request that a minimum of minnodes nodes be allocated to this job. A maximum node count may also be specified with maxnodes. If only one number is specified, this is used as both the minimum and maximum node count.
- -n, --ntasks=<number>
sbatch does not launch tasks, it requests an allocation of resources and submits a batch script. This option advises the Slurm controller that job steps run within the allocation will launch a maximum of number tasks and to provide for sufficient resources. The default is one task per node
- --mem=<MB>
Specify the real memory required per node in MegaBytes.
- -c, --cpus-per-task=<ncpus>
Advise the Slurm controller that ensuing job steps will require ncpus number of processors per task. Without this option, the controller will just try to allocate one processor per task.
- -p, --partition=<partition_names>
Request a specific partition for the resource allocation. Available partitions are: express(max 2 hrs), short(max 12 hrs), medium(max 50 hrs), long(max 150 hrs), sinteractive(0-2 hrs)
Submitting Jobs
Batch Jobs are submitted on Cheaha by using the "sbatch" command. The full manual for sbtach is available by running the following command
man sbatch
Job Script File Format
To submit a job to the queuing systems, you will first define your job in a script (a text file) and then submit that script to the queuing system.
The script file needs to be formatted as a UNIX file, not a Windows or Mac text file. In geek speak, this means that the end of line (EOL) character should be a line feed (LF) rather than a carriage return line feed (CRLF) for Windows or carriage return (CR) for Mac.
If you submit a job script formatted as a Windows or Mac text file, your job will likely fail with misleading messages, for example that the path specified does not exist.
Windows Notepad does not have the ability to save files using the UNIX file format. Do NOT use Notepad to create files intended for use on the clusters. Instead use one of the alternative text editors listed in the following section.
Converting Files to UNIX Format
Dos2Unix Method
The lines below that begin with $ are commands, the $ represents the command prompt and should not be typed!
The dos2unix program can be used to convert Windows text files to UNIX files with a simple command. After you have copied the file to your home directory on the cluster, you can identify that the file is a Windows file by executing the following (Windows uses CR LF as the line terminator, where UNIX uses only LF and Mac uses only CR):
$ file testfile.txt testfile.txt: ASCII text, with CRLF line terminators
Now, convert the file to UNIX
$ dos2unix testfile.txt dos2unix: converting file testfile.txt to UNIX format ...
Verify the conversion using the file command
$ file testfile.txt testfile.txt: ASCII text
Alternative Windows Text Editors
There are many good text editors available for Windows that have the capability to save files using the UNIX file format. Here are a few:
- [Geany] is an excellent free text editor for Windows and Linux that supports Windows, UNIX and Mac file formats, syntax highlighting and many programming features. To convert from Windows to UNIX click Document click Set Line Endings and then Convert and Set to LF (Unix)
- [Notepad++] is a great free Windows text editor that supports Windows, UNIX and Mac file formats, syntax highlighting and many programming features. To convert from Windows to UNIX click Format and then click Convert to UNIX Format
- [TextPad] is another excellent Windows text editor. TextPad is not free, however.
Example Batch Job Script
A shared cluster environment like Cheaha uses a job scheduler to run tasks on the cluster to provide optimal resource sharing among users. Cheaha uses a job scheduling system call Slurm to schedule and manage jobs. A user needs to tell Slurm about resource requirements (e.g. CPU, memory) so that it can schedule jobs effectively. These resource requirements along with actual application code can be specified in a single file commonly referred as 'Job Script/File'. Following is a simple job script that prints job number and hostname.
Note:Jobs must request the appropriate partition (ex: --partition=short) to satisfy the jobs resource request (maximum runtime, number of compute nodes, etc...)
#!/bin/bash # #SBATCH --job-name=test #SBATCH --output=res.txt #SBATCH --ntasks=1 #SBATCH --partition=express #SBATCH --time=10:00 #SBATCH --mem-per-cpu=100 #SBATCH --mail-type=FAIL #SBATCH --mail-user=YOUR_EMAIL_ADDRESS srun hostname srun sleep 60
Lines starting with '#SBATCH' have a special meaning in the Slurm world. Slurm specific configuration options are specified after the '#SBATCH' characters. Above configuration options are useful for most job scripts and for additional configuration options refer to Slurm commands manual. A job script is submitted to the cluster using Slurm specific commands. There are many commands available, but following three commands are the most common:
- sbatch - to submit job
- scancel - to delete job
- squeue - to view job status
We can submit above job script using sbatch command:
$ sbatch HelloCheaha.sh Submitted batch job 52707
When the job script is submitted, Slurm queues it up and assigns it a job number (e.g. 52707 in above example). The job number is available inside job script using environment variable $JOB_ID. This variable can be used inside job script to create job related directory structure or file names.
Interactive Resources
Login Node (the host that you connected to when you setup the SSH connection to Cheaha) is supposed to be used for submitting jobs and/or lighter prep work required for the job scripts. Do not run heavy computations on the login node. If you have a heavier workload to prepare for a batch job (eg. compiling code or other manipulations of data) or your compute application requires interactive control, you should request a dedicated interactive node for this work.
Interactive resources are requested by submitting an "interactive" job to the scheduler. Interactive jobs will provide you a command line on a compute resource that you can use just like you would the command line on the login node. The difference is that the scheduler has dedicated the requested resources to your job and you can run your interactive commands without having to worry about impacting other users on the login node.
Interactive jobs, that can be run on command line, are requested with the srun command.
srun --ntasks=1 --cpus-per-task=4 --mem-per-cpu=4096 --time=08:00:00 --partition=medium --job-name=JOB_NAME --pty /bin/bash
This command requests for 4 cores (--cpus-per-task) for a single task (--ntasks) with each cpu requesting size 4GB of RAM (--mem-per-cpu) for 8 hrs (--time).
More advanced interactive scenarios to support graphical applications are available using VNC or X11 tunneling X-Win32 2014 for Windows
Interactive jobs that requires running a graphical application, are requested with the sinteractive command, via Terminal on your VNC window.
sinteractive --ntasks=1 --cpus-per-task=4 --mem-per-cpu=4096 --time=08:00:00 --partition=medium --job-name=JOB_NAME
Please note, sinteractive starts your shell in a screen session. Screen is a terminal emulator that is designed to make it possible to detach and reattach a session. This feature can mostly be ignored. If you application uses `ctrl-a` as a special command sequence (e.g. Emacs), however, you may find the application doesn't receive this special character. When using screen, you need to type `ctrl-a a` (ctrl-a followed by a single "a" key press) to send a ctrl-a to your application. Screen uses ctrl-a as it's own command character, so this special sequence issues the command to screen to "send ctrl-a to my app". Learn more about screen from it's documentation.
Storage
Privacy
Do not store sensitive information on this filesystem. It is not encrypted. Note that your data will be stored on the cluster filesystem, and while not accessible to ordinary users, it could be accessible to the cluster administrator(s).
File and Directory Permissions
The default permissions for all user data storage locations described below are as follows. In these descriptions, the "$USER" variable should be replaced with the user's account name string:
- /home/$USER - the owner ($USER) of the directory can read, write/delete, and list files. No other users or groups have permissions to this directory.
- /data/user/$USER - the owner ($USER) of the directory can read, write/delete, and list files. No other users or groups have permissions to this directory.
- /scratch/$USER - the owner ($USER) of the directory can read, write/delete, and list files. No other users or groups have permissions to this directory.
- /data/projects/<projectname> - a PI can request project space for their lab or specific collaborations. The project directory is created with the PI/requestor as the user-owner and a dedicated collaboration group as the group-owner. The PI and all members of the dedicated collaboration group have can read, write/delete, and list files. No privileges are granted to other users of the system. Additional controls can be implemented via access control lists (ACLs). The PI/requestor can modify the ACLs to allow additional access to specific users.
These permissions are the default configuration. While it is possible to modify these permissions or change the group owner of a file to any group to which a user belongs, users are encouraged to work within the default configuration and contact support@listserv.uab.edu if the default permissions are not adequate. Setting up a collaboration group and associated project directory can address most collaboration need while keep data access restricted to the minimum necessary users for the collaboration.
Additional background on Linux file system permissions can be found here:
- https://its.unc.edu/research-computing/techdocs/how-to-use-unix-and-linux-file-permissions/
- https://www.rc.fas.harvard.edu/resources/documentation/linux/unix-permissions/
- https://hpc.nih.gov/storage/permissions.html
No Automatic Backups
There is no automatic back up of any user data on the cluster in home, data, or scratch. At this time, all user data back up processes are defined and managed by each user and/or lab. Given that data backup demands vary widely between different users, groups, and research domains, this approach enables those who are most familiar with the data to make appropriate decisions based on their specific needs.
For example, if a group is working with a large shared data set that is a local copy of a data set maintained authoritatively at a national data bank, maintaining a local backup is unlikely to be a productive use of limited storage resources, since this data could potentially be restored from the authoritative source. If, however, you are maintaining a unique source of data of which yours is the only copy, then maintaining a backup is critical if you value that data set. It's worth noting that while this "uniqueness" criteria may not apply to the data you analyze, it may readily apply to the codes that define your analysis pipelines.
An often recommended backup policy is the 3-2-1 rule: maintain three copies of data, on two different media, with one copy off-site. You can read more about the 3-2-1 rule here. In the case of your application codes, using revision control tools during development provides an easy way to maintain a second copy, makes for a good software development process, and can help achieve reproducible research goals.
Please review the data storage options provided by UAB IT for maintaining copies of your data. In choosing among these options, you should also be aware of UAB's data classification rules and requirements for security requirements for sensitive and restricted data storage. Given the importance of backup, Research Computing continues to explore options to facilitate data backup workflows from the cluster. Please contact us if you have questions or would like to discuss specific data backup scenarios.
A good guide for thinking about your backup strategy might be: "If you aren't managing a data back up process, then you have no backup data."
Home directories
Your home directory on Cheaha is NFS-mounted to the compute nodes as /home/$USER or $HOME. It is acceptable to use your home directory as a location to store job scripts and custom code. You are responsible for keeping your home directory under 10GB in size!
The home directory must not be used to store large amounts of data. Please use $USER_SCRATCH for actively used data sets and $USER_DATA for storage of non scratch data.
Scratch
Research Computing policy requires that all bulky input and output must be located on the scratch space. The home directory is intended to store your job scripts, log files, libraries and other supporting files.
Important Information:
- Scratch space (network and local) is not backed up.
- Research Computing expects each user to keep their scratch areas clean. The cluster scratch area are not to be used for archiving data.
Cheaha has two types of scratch space, network mounted and local.
- Network scratch ($USER_SCRATCH) is available on the login node and each compute node. This storage is a GPFS high performance file system providing roughly 4.7PB of usable storage. This should be your jobs primary working directory, unless the job would benefit from local scratch (see below).
- Local scratch is physically located on each compute node and is not accessible to the other nodes (including the login node). This space is useful if the job performs a lot of file I/O. Most of the jobs that run on our clusters do not fall into this category. Because the local scratch is inaccessible outside the job, it is important to note that you must move any data between local scratch to your network accessible scratch within your job. For example, step 1 in the job could be to copy the input from $USER_SCRATCH to ${USER_SCRATCH}, step 2 code execution, step 3 move the data back to $USER_SCRATCH.
Network Scratch
Network scratch is available using the environment variable $USER_SCRATCH or directly by /data/scratch/$USER
It is advisable to use the environment variable whenever possible rather than the hard coded path.
Local Scratch
Each compute node has a local scratch directory that is accessible via the variable $LOCAL_SCRATCH. If your job performs a lot of file I/O, the job should use $LOCAL_SCRATCH rather than $USER_SCRATCH to prevent bogging down the network scratch file system. The amount of scratch space available is approximately 800GB.
The $LOCAL_SCRATCH is a special temporary directory and it's important to note that this directory is deleted when the job completes, so the job script has to move the results to $USER_SCRATCH or other location prior to the job exiting.
Note that $LOCAL_SCRATCH is only useful for jobs in which all processes run on the same compute node, so MPI jobs are not candidates for this solution.
The following is an array job example that uses $LOCAL_SCRATCH by transferring the inputs into $LOCAL_SCRATCH at the beginning of the script and the result out of $LOCAL_SCRATCH at the end of the script.
#!/bin/bash #SBATCH --array=1-10 #SBATCH --share #SBATCH --partition=express # # Name your job to make it easier for you to track # #SBATCH --job-name=R_array_job # # Set your error and output files # #SBATCH --error=R_array_job.err #SBATCH --output=R_array_job.out #SBATCH --ntasks=1 # # Tell the scheduler only need 10 minutes and the appropriate partition # #SBATCH --time=00:10:00 #SBATCH --mem-per-cpu=256 # # Set your email address and request notification when you job is complete or if it fails # #SBATCH --mail-type=FAIL #SBATCH --mail-user=YOUR_EMAIL_ADDRESS module load R/3.2.0-goolf-1.7.20 echo "TMPDIR: $LOCAL_SCRATCH" cd $LOCAL_SCRATCH # Create a working directory under the special scheduler local scratch directory # using the array job's taskID mdkir $SLURM_ARRAY_TASK_ID cd $SLURM_ARRAY_TASK_ID # Next copy the input data to the local scratch echo "Copying input data from network scratch to $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID - $(date) # The input data in this case has a numerical file extension that # matches $SLURM_ARRAY_TASK_ID cp -a $USER_SCRATCH/GeneData/INP*.$SLURM_ARRAY_TASK_ID ./ echo "copied input data from network scratch to $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID - $(date) someapp -S 1 -D 10 -i INP*.$SLURM_ARRAY_TASK_ID -o geneapp.out.$SLURM_ARRAY_TASK_ID # Lastly copy the results back to network scratch echo "Copying results from local $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID to network - $(date) cp -a geneapp.out.$SLURM_ARRAY_TASK_ID $USER_SCRATCH/GeneData/ echo "Copied results from local $LOCAL_SCRATCH/$SLURM_ARRAY_TASK_ID to network - $(date)
Project Storage
Cheaha has a location where shared data can be stored called $SHARE_PROJECT. As with user scratch, this area is not backed up!
This is helpful if a team of researchers must access the same data. Please open a help desk ticket to request a project directory under $SHARE_PROJECT.
Uploading Data
Do not store sensitive information on this filesystem. It is not encrypted. Note that your data will be stored on the cluster filesystem, and while not accessible to ordinary users, it could be accessible to the cluster administrator(s). Data can be moved onto the cluster (pushed) from a remote client (ie. you desktop) via SCP or SFTP. Data can also be downloaded to the cluster (pulled) by issuing transfer commands once you are logged into the cluster. Common transfer methods are `wget <URL>`, FTP, or SCP, and depend on how the data is made available from the data provider.
Large data sets should be staged directly to your $USER_SCRATCH directory so as not to fill up $HOME. If you are working on a data set shared with multiple users, it's preferable to request space in $SHARE_PROJECT rather than duplicating the data for each user.
Environment Modules
Environment Modules is installed on Cheaha and should be used when constructing your job scripts if an applicable module file exists. Using the module command you can easily configure your environment for specific software packages without having to know the specific environment variables and values to set. Modules allows you to dynamically configure your environment without having to logout / login for the changes to take affect.
If you find that specific software does not have a module, please submit a helpdesk ticket to request the module.
- Cheaha supports bash completion for the module command. For example, type 'module' and press the TAB key twice to see a list of options:
module TAB TAB add display initlist keyword refresh switch use apropos help initprepend list rm unload whatis avail initadd initrm load show unuse clear initclear initswitch purge swap update
- To see the list of available modulefiles on the cluster, run the module avail command (note the example list below may not be complete!) or module load followed by two tab key presses:
module avail ----------------------------------------------------------------------------------------- /cm/shared/modulefiles ----------------------------------------------------------------------------------------- acml/gcc/64/5.3.1 acml/open64-int64/mp/fma4/5.3.1 fftw2/openmpi/gcc/64/float/2.1.5 intel-cluster-runtime/ia32/3.8 netcdf/gcc/64/4.3.3.1 acml/gcc/fma4/5.3.1 blacs/openmpi/gcc/64/1.1patch03 fftw2/openmpi/open64/64/double/2.1.5 intel-cluster-runtime/intel64/3.8 netcdf/open64/64/4.3.3.1 acml/gcc/mp/64/5.3.1 blacs/openmpi/open64/64/1.1patch03 fftw2/openmpi/open64/64/float/2.1.5 intel-cluster-runtime/mic/3.8 netperf/2.7.0 acml/gcc/mp/fma4/5.3.1 blas/gcc/64/3.6.0 fftw3/openmpi/gcc/64/3.3.4 intel-tbb-oss/ia32/44_20160526oss open64/4.5.2.1 acml/gcc-int64/64/5.3.1 blas/open64/64/3.6.0 fftw3/openmpi/open64/64/3.3.4 intel-tbb-oss/intel64/44_20160526oss openblas/dynamic/0.2.15 acml/gcc-int64/fma4/5.3.1 bonnie++/1.97.1 gdb/7.9 iozone/3_434 openmpi/gcc/64/1.10.1 acml/gcc-int64/mp/64/5.3.1 cmgui/7.2 globalarrays/openmpi/gcc/64/5.4 lapack/gcc/64/3.6.0 openmpi/open64/64/1.10.1 acml/gcc-int64/mp/fma4/5.3.1 cuda75/blas/7.5.18 globalarrays/openmpi/open64/64/5.4 lapack/open64/64/3.6.0 pbspro/13.0.2.153173 acml/open64/64/5.3.1 cuda75/fft/7.5.18 hdf5/1.6.10 mpich/ge/gcc/64/3.2 puppet/3.8.4 acml/open64/fma4/5.3.1 cuda75/gdk/352.79 hdf5_18/1.8.16 mpich/ge/open64/64/3.2 rc-base acml/open64/mp/64/5.3.1 cuda75/nsight/7.5.18 hpl/2.1 mpiexec/0.84_432 scalapack/mvapich2/gcc/64/2.0.2 acml/open64/mp/fma4/5.3.1 cuda75/profiler/7.5.18 hwloc/1.10.1 mvapich/gcc/64/1.2rc1 scalapack/openmpi/gcc/64/2.0.2 acml/open64-int64/64/5.3.1 cuda75/toolkit/7.5.18 intel/compiler/32/15.0/2015.5.223 mvapich/open64/64/1.2rc1 sge/2011.11p1 acml/open64-int64/fma4/5.3.1 default-environment intel/compiler/64/15.0/2015.5.223 mvapich2/gcc/64/2.2b slurm/15.08.6 acml/open64-int64/mp/64/5.3.1 fftw2/openmpi/gcc/64/double/2.1.5 intel-cluster-checker/2.2.2 mvapich2/open64/64/2.2b torque/6.0.0.1 ---------------------------------------------------------------------------------------- /share/apps/modulefiles ----------------------------------------------------------------------------------------- rc/BrainSuite/15b rc/freesurfer/freesurfer-5.3.0 rc/intel/compiler/64/ps_2016/2016.0.047 rc/matlab/R2015a rc/SAS/v9.4 rc/cmg/2012.116.G rc/gromacs-intel/5.1.1 rc/Mathematica/10.3 rc/matlab/R2015b rc/dsistudio/dsistudio-20151020 rc/gtool/0.7.5 rc/matlab/R2012a rc/MRIConvert/2.0.8 --------------------------------------------------------------------------------------- /share/apps/rc/modules/all --------------------------------------------------------------------------------------- AFNI/linux_openmp_64-goolf-1.7.20-20160616 gperf/3.0.4-intel-2016a MVAPICH2/2.2b-GCC-4.9.3-2.25 Amber/14-intel-2016a-AmberTools-15-patchlevel-13-13 grep/2.15-goolf-1.4.10 NASM/2.11.06-goolf-1.7.20 annovar/2016Feb01-foss-2015b-Perl-5.22.1 GROMACS/5.0.5-intel-2015b-hybrid NASM/2.11.08-foss-2015b ant/1.9.6-Java-1.7.0_80 GSL/1.16-goolf-1.7.20 NASM/2.11.08-intel-2016a APBS/1.4-linux-static-x86_64 GSL/1.16-intel-2015b NASM/2.12.02-foss-2016a ASHS/rev103_20140612 GSL/2.1-foss-2015b NASM/2.12.02-intel-2015b Aspera-Connect/3.6.1 gtool/0.7.5_linux_x86_64 NASM/2.12.02-intel-2016a ATLAS/3.10.1-gompi-1.5.12-LAPACK-3.4.2 guile/1.8.8-GNU-4.9.3-2.25 ncurses/5.9-foss-2015b Autoconf/2.69-foss-2016a HAPGEN2/2.2.0 ncurses/5.9-GCC-4.8.4 Autoconf/2.69-GCC-4.8.4 HarfBuzz/1.2.7-intel-2016a ncurses/5.9-GNU-4.9.3-2.25 Autoconf/2.69-GNU-4.9.3-2.25 HDF5/1.8.15-patch1-intel-2015b ncurses/5.9-goolf-1.4.10 . . . .
Some software packages have multiple module files, for example:
- GCC/4.7.2
- GCC/4.8.1
- GCC/4.8.2
- GCC/4.8.4
- GCC/4.9.2
- GCC/4.9.3
- GCC/4.9.3-2.25
In this case, the GCC module will always load the latest version, so loading this module is equivalent to loading GCC/4.9.3-2.25. If you always want to use the latest version, use this approach. If you want use a specific version, use the module file containing the appropriate version number.
Some modules, when loaded, will actually load other modules. For example, the GROMACS/5.0.5-intel-2015b-hybrid module will also load intel/2015b and other related tools.
- To load a module, ex: for a GROMACS job, use the following module load command in your job script:
module load GROMACS/5.0.5-intel-2015b-hybrid
- To see a list of the modules that you currently have loaded use the module list command
module list Currently Loaded Modulefiles: 1) slurm/15.08.6 9) impi/5.0.3.048-iccifort-2015.3.187-GNU-4.9.3-2.25 17) Tcl/8.6.3-intel-2015b 2) rc-base 10) iimpi/7.3.5-GNU-4.9.3-2.25 18) SQLite/3.8.8.1-intel-2015b 3) GCC/4.9.3-binutils-2.25 11) imkl/11.2.3.187-iimpi-7.3.5-GNU-4.9.3-2.25 19) Tk/8.6.3-intel-2015b-no-X11 4) binutils/2.25-GCC-4.9.3-binutils-2.25 12) intel/2015b 20) Python/2.7.9-intel-2015b 5) GNU/4.9.3-2.25 13) bzip2/1.0.6-intel-2015b 21) Boost/1.58.0-intel-2015b-Python-2.7.9 6) icc/2015.3.187-GNU-4.9.3-2.25 14) zlib/1.2.8-intel-2015b 22) GROMACS/5.0.5-intel-2015b-hybrid 7) ifort/2015.3.187-GNU-4.9.3-2.25 15) ncurses/5.9-intel-2015b 8) iccifort/2015.3.187-GNU-4.9.3-2.25 16) libreadline/6.3-intel-2015b
- A module can be removed from your environment by using the module unload command:
module unload GROMACS/5.0.5-intel-2015b-hybrid
- The definition of a module can also be viewed using the module show command, revealing what a specific module will do to your environment:
module show GROMACS/5.0.5-intel-2015b-hybrid ------------------------------------------------------------------- /share/apps/rc/modules/all/GROMACS/5.0.5-intel-2015b-hybrid: module-whatis GROMACS is a versatile package to perform molecular dynamics, i.e. simulate the Newtonian equations of motion for systems with hundreds to millions of particles. - Homepage: http://www.gromacs.org conflict GROMACS prepend-path CPATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/include prepend-path LD_LIBRARY_PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/lib64 prepend-path LIBRARY_PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/lib64 prepend-path MANPATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/share/man prepend-path PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/bin prepend-path PKG_CONFIG_PATH /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/lib64/pkgconfig setenv EBROOTGROMACS /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid setenv EBVERSIONGROMACS 5.0.5 setenv EBDEVELGROMACS /share/apps/rc/software/GROMACS/5.0.5-intel-2015b-hybrid/easybuild/GROMACS-5.0.5-intel-2015b-hybrid-easybuild-devel -------------------------------------------------------------------
Error Using Modules from a Job Script
If you are using modules and the command your job executes runs fine from the command line but fails when you run it from the job, you may be having an issue with the script initialization. If you see this error in your job error output file
-bash: module: line 1: syntax error: unexpected end of file -bash: error importing function definition for `BASH_FUNC_module'
Add the command `unset module` before calling your module files. The -V job argument will cause a conflict with the module function used in your script.
Sample Job Scripts
The following are sample job scripts, please be careful to edit these for your environment (i.e. replace YOUR_EMAIL_ADDRESS with your real email address), set the h_rt to an appropriate runtime limit and modify the job name and any other parameters.
Hello World is the classic example used throughout programming. We don't want to buck the system, so we'll use it as well to demonstrate jobs submission with one minor variation: our hello world will send us a greeting using the name of whatever machine it runs on. For example, when run on the Cheaha login node, it would print "Hello from login001".
Hello World (serial)
A serial job is one that can run independently of other commands, ie. it doesn't depend on the data from other jobs running simultaneously. You can run many serial jobs in any order. This is a common solution to processing lots of data when each command works on a single piece of data. For example, running the same conversion on 100s of images.
Here we show how to create job script for one simple command. Running more than one command just requires submitting more jobs.
- Create your hello world application. Run this command to create a script, turn it into to a command, and run the command (just copy and past the following on to the command line).
1. Create the file:
$ vim helloworld.sh
2. Write into "helloworld.sh" file (To write in vim editor: press shift + I )
#!/bin/bash echo Hello from `hostname`
3. Save the file by pressing the esc key, type the following
:wq
4. Need to give permission the "helloworld.sh" file
$ chmod +x helloworld.sh
- Create the Slurm job script that will request 256 MB RAM and a maximum runtime of 10 minutes.
1. Create the JOB file:
$ vim helloworld.job
2. Write into "helloworld.job" file (To write in vim editor: press shift + I )
#!/bin/bash #SBATCH --share #SBATCH --partition=express # # Name your job to make it easier for you to track # #SBATCH --job-name=helloworld # # Set your error and output files # #SBATCH --error=helloworld.err #SBATCH --output=helloworld.out #SBATCH --ntasks=1 # # Tell the scheduler only need 10 minutes # #SBATCH --time=00:10:00 #SBATCH --mem-per-cpu=256 # # Set your email address and request notification when you job is complete or if it fails # #SBATCH --mail-type=FAIL #SBATCH --mail-user=$USER@uab.edu ./helloworld.sh
3. Save the file by pressing the esc key, type the following
:wq
- Submit the job to Slurm scheduler and check the status using squeue
$ sbatch helloworld.job Submitted batch job 52888
- When the job completes, you should have output files named helloworld.out and helloworld.err
$ cat helloworld.out Hello from c0003
Hello World (parallel with MPI)
MPI is used to coordinate the activity of many computations occurring in parallel. It is commonly used in simulation software for molecular dynamics, fluid dynamics, and similar domains where there is significant communication (data) exchanged between cooperating process.
Here is a simple parallel Slurm job script for running commands the rely on MPI. This example also includes the example of compiling the code and submitting the job script to the Slurm scheduler.
- First, create a directory for the Hello World jobs
$ mkdir -p ~/jobs/helloworld $ cd ~/jobs/helloworld
- Create the Hello World code written in C (this example of MPI enabled Hello World includes a 3 minute sleep to ensure the job runs for several minutes, a normal hello world example would run in a matter of seconds).
$ vi helloworld-mpi.c
#include <stdio.h>
#include <mpi.h>
main(int argc, char **argv)
{
int rank, size;
int i, j;
float f;
MPI_Init(&argc,&argv);
MPI_Comm_rank(MPI_COMM_WORLD, &rank);
MPI_Comm_size(MPI_COMM_WORLD, &size);
printf("Hello World from process %d of %d.\n", rank, size);
sleep(180);
for (j=0; j<=100000; j++)
for(i=0; i<=100000; i++)
f=i*2.718281828*i+i+i*3.141592654;
MPI_Finalize();
}
- Compile the code, first purging any modules you may have loaded followed by loading the module for OpenMPI GNU. The mpicc command will compile the code and produce a binary named helloworld_gnu_openmpi
$ module purge $ module load DefaultModules $ module load OpenMPI/4.0.1-GCC-8.3.0-2.32 $ mpicc helloworld-mpi.c -o helloworld_gnu_openmpi
- Create the Slurm job script that will request 8 cpu slots and a maximum runtime of 10 minutes
$ vi helloworld.job
#!/bin/bash #SBATCH --share #SBATCH --partition=express # # Name your job to make it easier for you to track # #SBATCH --job-name=helloworld_mpi # # Set your error and output files # #SBATCH --error=helloworld_mpi.err #SBATCH --output=helloworld_mpi.out #SBATCH --ntasks=8 # # Tell the scheduler only need 10 minutes # #SBATCH --time=00:10:00 #SBATCH --mem-per-cpu=256 # # Set your email address and request notification when you job is complete or if it fails # #SBATCH --mail-type=FAIL #SBATCH --mail-user=YOUR_EMAIL_ADDRESS module load OpenMPI/1.8.8-GNU-4.9.3-2.25 mpirun -np $SLURM_NTASKS helloworld_gnu_openmpi
- Submit the job to Slurm scheduler and check the status using squeue -u $USER
$ sbatch helloworld.job
Submitted batch job 52893
$ squeue -u BLAZERID
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
52893 express hellowor BLAZERID R 2:07 2 c[0005-0006]
- When the job completes, you should have output files named helloworld_mpi.out and helloworld_mpi.err
$ cat helloworld_mpi.out Hello World from process 1 of 8. Hello World from process 3 of 8. Hello World from process 4 of 8. Hello World from process 7 of 8. Hello World from process 5 of 8. Hello World from process 6 of 8. Hello World from process 0 of 8. Hello World from process 2 of 8.
Hello World (serial) -- revisited
The job submit scripts (sbatch scripts) are actually bash shell scripts in their own right. The reason for using the funky #SBATCH prefix in the scripts is so that bash interprets any such line as a comment and won't execute it. Because the # character starts a comment in bash, we can weave the Slurm scheduler directives (the #SBATCH lines) into standard bash scripts. This lets us build scripts that we can execute locally and then easily run the same script to on a cluster node by calling it with sbatch. This can be used to our advantage to create a more fluid experience in moving between development and production job runs.
The following example is a simple variation on the serial job above. All we will do is convert our Slurm job script into a command called helloworld that calls the helloworld.sh command.
If the first line of a file is #!/bin/bash and that file is executable, the shell will automatically run the command as if were any other system command, eg. ls. That is, the ".sh" extension on our HelloWorld.sh script is completely optional and is only meaningful to the user.
Copy the serial helloworld.job script to a new file, add a the special #!/bin/bash as the first line, and make it executable with the following command (note: those are single quotes in the echo command):
echo '#!/bin/bash' | cat helloworld.job > helloworld ; chmod +x helloworld
Our sbatch script has now become a regular command. We can now execute the command with the simple prefix "./helloworld", which means "execute this file in the current directory":
./helloworld Hello from login001
Or if we want to run the command on a compute node, replace the "./" prefix with "sbatch ":
$ sbatch helloworld Submitted batch job 53001
And when the cluster run is complete you can look at the content of the output:
$ $ cat helloworld.out Hello from c0003
You can use this approach of treating you sbatch files as command wrappers to build a collection of commands that can be executed locally or via sbatch. The other examples can be restructured similarly.
To avoid having to use the "./" prefix, just add the current directory to your PATH. Also, if you plan to do heavy development using this feature on the cluster, please be sure to run sinteractive first so you don't load the login node with your development work.
Gromacs
#!/bin/bash
#SBATCH --partition=short
#
# Name your job to make it easier for you to track
#
#SBATCH --job-name=test_gromacs
#
# Set your error and output files
#
#SBATCH --error=test_gromacs.err
#SBATCH --output=test_gromacs.out
#SBATCH --ntasks=8
#
# Tell the scheduler only need 10 minutes
#
#SBATCH --time=10:00:00
#SBATCH --mem-per-cpu=2048
#
# Set your email address and request notification when you job is complete or if it fails
#
#SBATCH --mail-type=FAIL
#SBATCH --mail-user=YOUR_EMAIL_ADDRESS
module load OpenMPI/1.8.8-GNU-4.9.3-2.25
module load GROMACS/5.0.5-intel-2015b-hybrid
# Change directory to the job working directory if not already there
cd ${USER_SCRATCH}/jobs/gromacs
# Single precision
MDRUN=mdrun_mpi
# Enter your tpr file over here
export MYFILE=example.tpr
mpirun -np SLURM_NTASKS $MDRUN -v -s $MYFILE -o $MYFILE -c $MYFILE -x $MYFILE -e $MYFILE -g ${MYFILE}.log
R (array job)
The following is an example job script that will use an array of 10 tasks (--array=1-10), each task has a max runtime of 2 hours and will use no more than 256 MB of RAM per task. Array's of tasks are useful when you have lots of simple jobs that work on their own separate files or a sub-set of the problem that can be selected by the array task index. For a more comprehensive introduction please see this tutorial.
Create a working directory and the job submission script
$ mkdir -p ~/jobs/ArrayExample $ cd ~/jobs/ArrayExample $ vi R-example-array.job
#!/bin/bash #SBATCH --array=1-10 #SBATCH --share #SBATCH --partition=express # # Name your job to make it easier for you to track # #SBATCH --job-name=R_array_job # # Set your error and output files # #SBATCH --error=R_array_job.err #SBATCH --output=R_array_job.out #SBATCH --ntasks=1 # # Tell the scheduler only need 10 minutes # #SBATCH --time=00:10:00 #SBATCH --mem-per-cpu=256 # # Set your email address and request notification when you job is complete or if it fails # #SBATCH --mail-type=FAIL #SBATCH --mail-user=YOUR_EMAIL_ADDRESS module load R/3.2.0-goolf-1.7.20 cd ~/jobs/ArrayExample/rep$SLURM_ARRAY_TASK_ID srun R CMD BATCH rscript.R
Submit the job to the Slurm scheduler and check the status of the job using the squeue command
$ sbatch R-example-array.job $ squeue -u $USER
Array Job Parameterization
Suppose you need to submit thousands of jobs. While you could do this in a for loop, the global limit on jobs in the SLURM queue is 10,000. The limit is in place for performance reasons and the jobs may be rejected with the following error message and an incomplete set of tasks.
sbatch: error: Slurm temporarily unable to accept job, sleeping and retrying
The preferred way to handle this scenario is to allow SLURM to schedule the jobs for you using the array flag in an sbatch script. This allows many jobs to be submitted as a single entry in the queue, letting SLURM handle the for loop and queueing. It is possible to reference the current loop index, or task id, as $SLURM_ARRAY_TASK_ID.
An example using $SLURM_ARRAY_TASK_ID to load input files and create output files is shown below. Suppose you have a short script called my_processing_script that needs to be run on 20,000 separate files. Suppose each instance only needs 1 cpu and 2 GB of RAM and finishes in 5 minutes. Submitting these files all at once won't work and at least half of them will be rejected by SLURM. Instead we can use the sbatch array flag. Note that some other useful flags have been omitted for brevity.
#! /bin/bash #SBATCH --partition=express #SBATCH --ntasks=1 #SBATCH --cpus-per-task=1 #SBATCH --mem-per-cpu=2G #SBATCH --array=1-20000%100 # This will run tasks 1 through 20000, with up to 100 at a time. # It is possible to provide any comma-separated list of intervals. # An example of a valid subset is --array=1,2,5-1000,3777,4995-5000%100 INPUT_FILE=$USER_DATA/input/file_$SLURM_ARRAY_TASK_ID.txt OUTPUT_FILE=$USER_DATA/output/file_$SLURM_ARRAY_TASK_ID.txt my_processing_script --input="$INPUT_FILE" --output="$OUTPUT_FILE"
GPU JOB
A Graphics processing unit (GPU) is a specialized electronic circuit designed to rapidly manipulate and alter memory to accelerate the creation of images in a frame buffer intended for output to a display device. Create a math.sh file as:
$vim math.sh
#!/bin/bash (e=5) echo $e (( e = e + 3 )) echo $e (( e=e+4 )) # -- spaces or no spaces, it doesn't matter echo $e
Give File permissions for script as follows:
$chmod +x math.sh
Create Job submission script file:
$vi math.job
#!/bin/bash #SBATCH --share #SBATCH --partition=pascalnodes #SBATCH --gres=gpu:1 # Name your job to make it easier for you to track # #SBATCH --job-name=math # # Set your error and output files # #SBATCH --error=math.err #SBATCH --output=math.out #SBATCH --ntasks=1 # # Tell the scheduler only need 10 minutes # #SBATCH --time=00:10:00 #SBATCH --mem-per-cpu=256 # # Set your email address and request notification when you job is complete or if it fails # #SBATCH --mail-type=FAIL #SBATCH --mail-user=$USER@uab.edu ./math.sh
Submitting batch script to Slurm scheduler
$sbatch math.job
We can also request GPU's on cluster as:
$sinteractive --ntasks=1 --time=00:10:00 --exclusive --partition=pascalnodes -N2 --gres=gpu:2
GPU Job (with MPI)
As mentioned above, MPI is used to coordinate the activity of many computations occurring in parallel. It is commonly used in simulation software for molecular dynamics, fluid dynamics, and similar domains where there is significant communication (data) exchanged between cooperating process.
An example of an GPU job with MPI can be found by visiting this link.
Be sure to request the appropiate amount of gpu resources for your job:
sinteractive --ntasks=8 --time=08:00:00 --exclusive --partition=pascalnodes -N2 --gres=gpu:4
Singularity Container
Singularity is designed so that you can use it within SLURM jobs and it does not violate security constraints on the cluster. Singularity was built keeping HPC in mind, i.e a shared environment. Using Singularity container with SLURM job script is very easy, as the containers run as a process on the host machine, just like any other command in a batch script. You just need to load Singularity in your job script and run the command via a singularity process. Here's an example job script below:
#!/bin/bash # #SBATCH --job-name=test-singularity #SBATCH --output=res.out #SBATCH --error=res.err # # Number of tasks needed for this job. Generally, used with MPI jobs #SBATCH --ntasks=1 #SBATCH --partition=express # # Time format = HH:MM:SS, DD-HH:MM:SS #SBATCH --time=10:00 # # Number of CPUs allocated to each task. #SBATCH --cpus-per-task=1 # # Mimimum memory required per allocated CPU in MegaBytes. #SBATCH --mem-per-cpu=100 # # Send mail to the email address when the job fails #SBATCH --mail-type=FAIL #SBATCH --mail-user=$USER@uab.edu #Set your environment here module load Singularity/2.5.2-GCC-5.4.0-2.26 #Run your singularity or any other commands here singularity exec -B /data/user/$USER /data/user/$USER/rc-training-sessions/neurodebian-neurodebian-master-latest.simg dcm2nii PATH_TO_YOUR_DICOM_FILES
For a more comprehensive introduction please see this tutorial.
Installed Software
A partial list of installed software with additional instructions for their use is available on the Cheaha Software page.