Galaxy DNA-Seq Tutorial: Difference between revisions
Jump to navigation
Jump to search
(Linking in WR data) |
|||
| Line 8: | Line 8: | ||
* Click the top box to select all 6 files | * Click the top box to select all 6 files | ||
* Select "import to current history" | * Select "import to current history" | ||
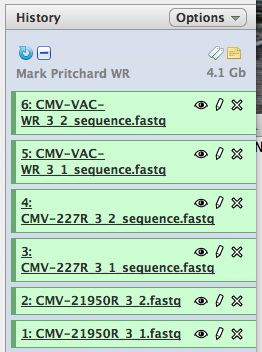
* Click on "Analyze Data" from the upper main menu. It should bring up the main page and your history pane should now look like the image below. | |||
[[File:HistoryPane.jpg]] | |||
* Notice that with just 3 viruses are already over 4 GB of data. | |||
* Click on the pencil to icon in one of the virus images to pull up the attributes, your screen should look a bit like this: | |||
== Formatting and Grooming Data == | == Formatting and Grooming Data == | ||
Revision as of 21:29, 13 September 2011
Galaxy DNA-Seq Tutorial
Linking to data
Link in the Mark Pritchard Vaccinia virus data set.
- Start with a blank history, there should be no numbered items on the right hand side of the pane. Otherwise create a new history.
- Select "Shared Data" from the top of the screen to bring up the Shared Data screen
- Select "Mark Pritchard Vaccinia WR" from the alphabetically sorted list
- Click the top box to select all 6 files
- Select "import to current history"
- Click on "Analyze Data" from the upper main menu. It should bring up the main page and your history pane should now look like the image below.
- Notice that with just 3 viruses are already over 4 GB of data.
- Click on the pencil to icon in one of the virus images to pull up the attributes, your screen should look a bit like this: