Galaxy DNA-Seq Tutorial: Difference between revisions
Jump to navigation
Jump to search
(SNPEff separate section) |
No edit summary |
||
| Line 73: | Line 73: | ||
* PATRIC, annotation sites | * PATRIC, annotation sites | ||
== Variant Summation == | == SNPEff and Variant Summation == | ||
Summarizing variants and effects of SNPs. | Summarizing variants and effects of SNPs. | ||
* Reference genome must be in SNPEff, need GFF3 file of your genome annotation | * Reference genome must be in SNPEff, need GFF3 file of your genome annotation | ||
| Line 79: | Line 79: | ||
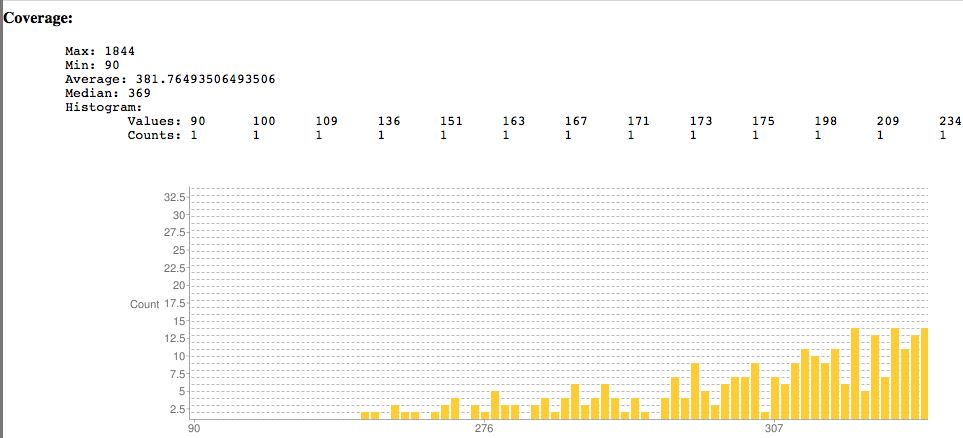
* Plots coverage, indel sizes | * Plots coverage, indel sizes | ||
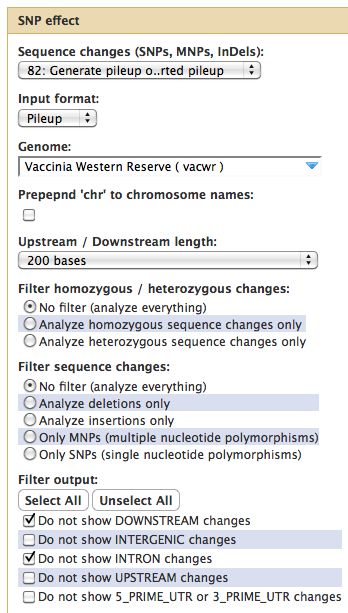
=== SNPEff === | === Running SNPEff === | ||
[[File:SNPEff.jpg]] | |||
=== Results from SNPEff | |||
* Only as good as your genome annotation file | |||
* Introns in viruses? | |||
[[File:SNPEffCoverage.jpg]] | |||
== De novo assembly (time permitting) == | == De novo assembly (time permitting) == | ||
Revision as of 04:20, 15 September 2011
Galaxy DNA-Seq Tutorial
Linking to data
Link in the Mark Pritchard Vaccinia virus data set.
- Start with a blank history, there should be no numbered items on the right hand side of the pane. Otherwise create a new history.
- Select "Shared Data" from the top of the screen to bring up the Shared Data screen
- Select "Mark Pritchard Vaccinia WR" from the alphabetically sorted list
- Click the top box to select all 6 files
- Select "import to current history"
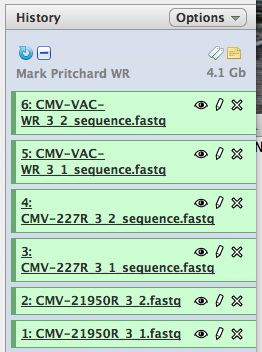
- Click on "Analyze Data" from the upper main menu. It should bring up the main page and your history pane should now look like the image below.
- Notice that with just 3 viruses are already over 4 GB of data.
Formatting and Grooming Data
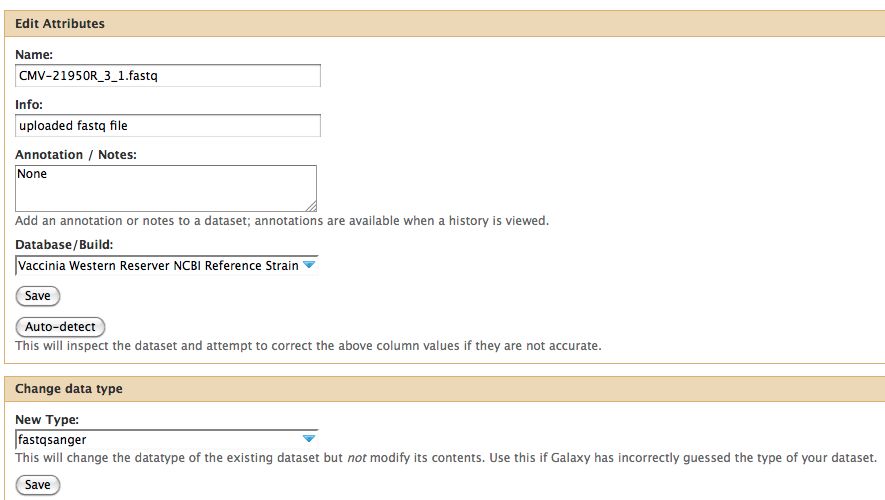
- Click on the pencil to icon in one of the virus images to pull up the attributes, your screen should look a bit like this:
- The important thing to notice is the data type. In Galaxy the expected data type of the galaxy tool must match EXACTLY with the data type in your history pane, otherwise the option to use that particular piece of data will not appear in the tool's drop down menu for data selection.
- There are multiple types of FastQ format, see the wikipedia article on FastQ for an idea. Galaxy requires that everything go into Sanger format to be used. If you know your data is in sanger format, select fastqsanger for your data type. If it is not in that format, select fastq and run the FastQ Groomer.
Assessing the quality of the data
We will use a number of different tools from the "NGS: QC and manipulation" drop down menu. Try processing the FastQ files with:
- FastQC
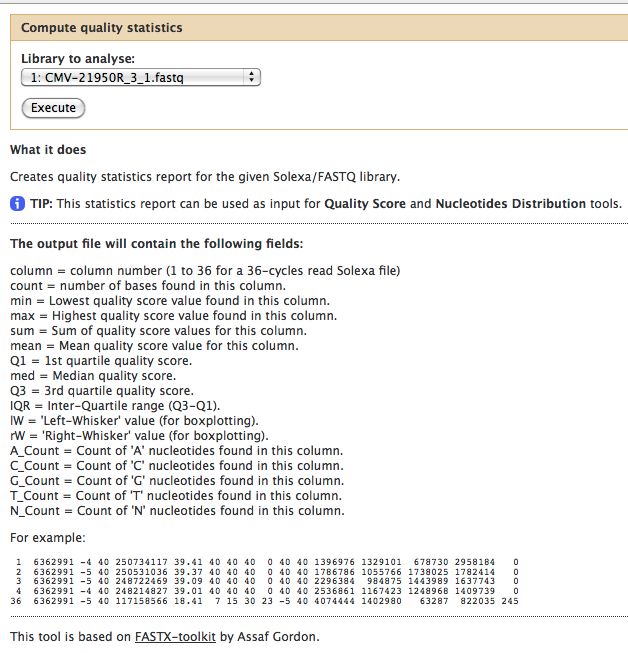
- Compute Quality Statistics
FastQC gives an attractive visual output and will flag potential problems.
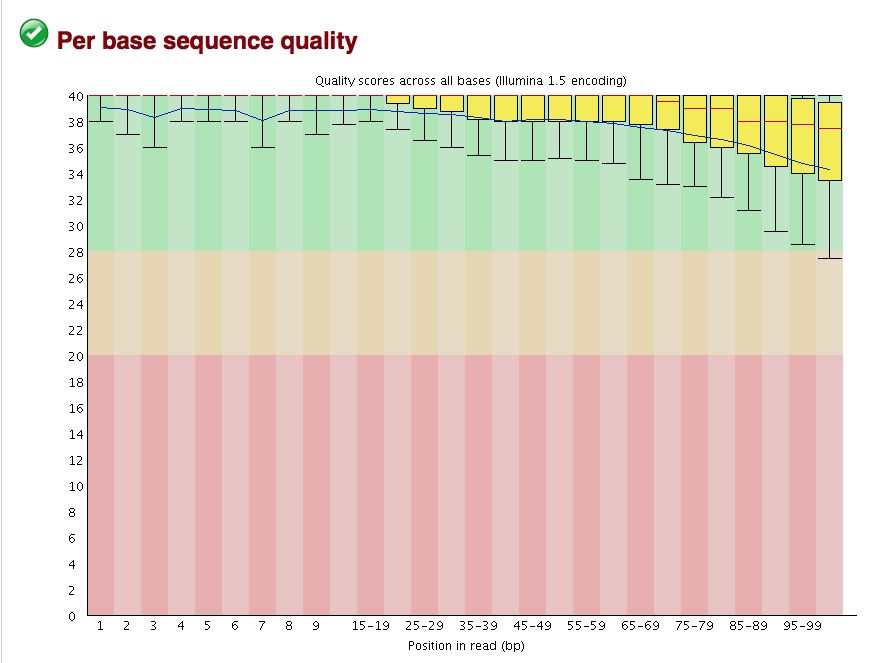
Take a look at the other areas that show up as quality issues. Can any of these be a problem for us?
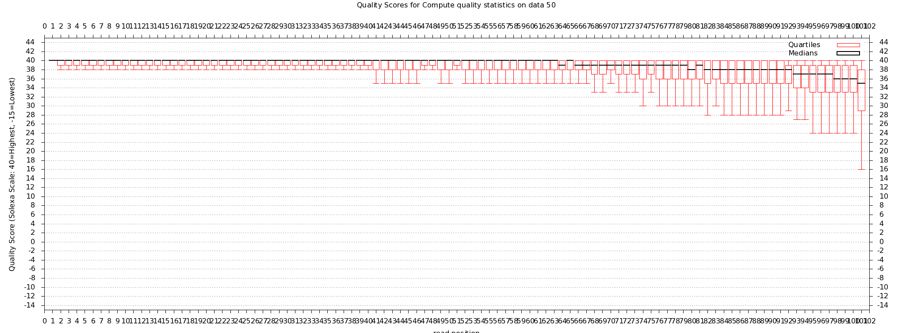
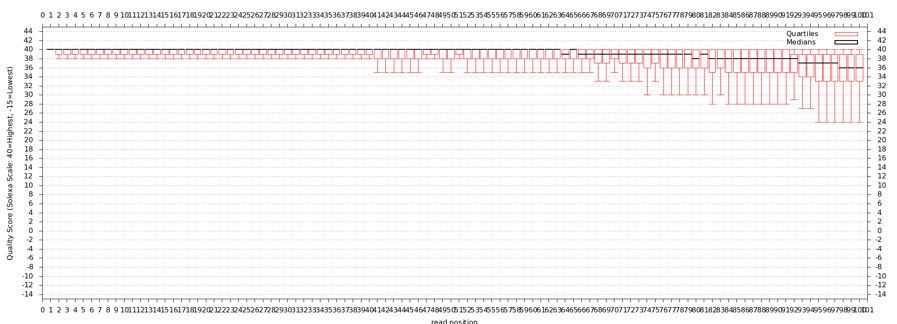
The quality statistics computing by the 2nd operation can be used for another box and whiskers plot to look at the read quality on a base by base basis. Select:
- "NGS: QC and manipulation" -> Draw Quality Score Boxplot
Performing cleanup
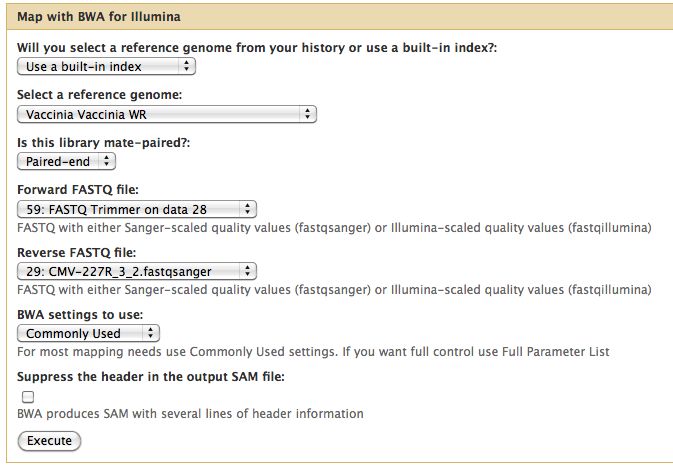
Short read alignment to reference genome using BWA
BAM Conversion
The output of many mapping programs is in SAM format and must be converted to the binary format. The conversion is lossless.
Variant Analysis
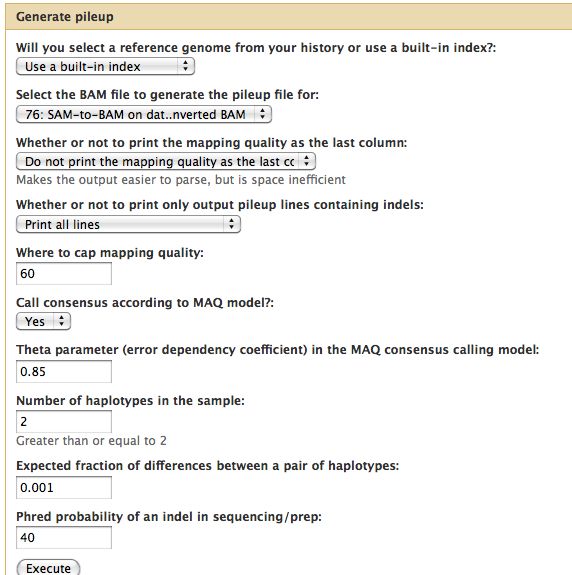
Samtools
- Essential toolset, performs a variety of functions including generation of a "pileup" file.
Other Variant Detection Approaches
- GATK - Emerging as the new best practise to call variants, not integrated into Galaxy just yet.
- SNPEff
- PATRIC, annotation sites
SNPEff and Variant Summation
Summarizing variants and effects of SNPs.
- Reference genome must be in SNPEff, need GFF3 file of your genome annotation
- SNPEff looks at heterozygous and homozygous SNPs, MNPs
- Plots coverage, indel sizes
Running SNPEff
=== Results from SNPEff
- Only as good as your genome annotation file
- Introns in viruses?